Discriminating Benign from Malignant Lung Diseases Using Plasma Glycosaminoglycans and Cell-Free DNA
- PMID: 39337265
- PMCID: PMC11431521
- DOI: 10.3390/ijms25189777
Discriminating Benign from Malignant Lung Diseases Using Plasma Glycosaminoglycans and Cell-Free DNA
Abstract
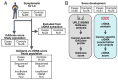
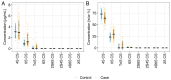
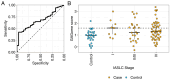
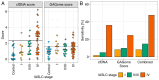
We aimed to investigate the use of free glycosaminoglycan profiles (GAGomes) and cfDNA in plasma to differentiate between lung cancer and benign lung disease, in a cohort of 113 patients initially suspected of lung cancer. GAGomes were analyzed in all samples using the MIRAM® Free Glycosaminoglycan Kit with ultra-high-performance liquid chromatography and electrospray ionization triple quadrupole mass spectrometry. In a subset of samples, cfDNA concentration and NGS-data was available. We detected two GAGome features, 0S chondroitin sulfate (CS), and 4S CS, with cancer-specific changes. Based on the observed GAGome changes, we devised a model to predict lung cancer. The model, named the GAGome score, could detect lung cancer with 41.2% sensitivity (95% CI: 9.2-54.2%) at 96.4% specificity (95% CI: 95.2-100.0%, n = 113). When we combined the GAGome score with a cfDNA-based model, the sensitivity increased from 42.6% (95% CI: 31.7-60.6%, cfDNA alone) to 70.5% (95% CI: 57.4-81.5%) at 95% specificity (95% CI: 75.1-100%, n = 74). Notably, the combined GAGome and cfDNA testing improved the sensitivity, compared to cfDNA alone, especially in ASCL stage I (55.6% vs 11.1%). Our findings show that plasma GAGome profiles can enhance cfDNA testing performance, highlighting the applicability of a multiomics approach in lung cancer diagnostics.
Keywords: GAGome; cfDNA; glycosaminoglycans; lung cancer; multiomics.
Conflict of interest statement
F. Gatto and J. Nielsen are shareholders in Elypta AB. F. Gatto and S. Bratulic are employed by Elypta AB. J. Nielsen is a board member at Elypta AB. Elypta AB has a commercial interest in part of the technology described in this study. The remaining authors have no conflicts to declare.
Figures
References
-
- Wang P., Song Q., Ren J., Zhang W., Wang Y., Zhou L., Wang D., Chen K., Jiang L., Zhang B., et al. Simultaneous analysis of mutations and methylations in circulating cell-free DNA for hepatocellular carcinoma detection. Sci. Transl. Med. 2022;14:eabp8704. doi: 10.1126/scitranslmed.abp8704. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

