Analysis of the Rice Raffinose Synthase (OsRS) Gene Family and Haplotype Diversity
- PMID: 39337301
- PMCID: PMC11432550
- DOI: 10.3390/ijms25189815
Analysis of the Rice Raffinose Synthase (OsRS) Gene Family and Haplotype Diversity
Abstract
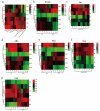
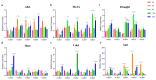
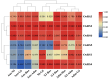
Based on the genome information of rice (Nipponbare), this study screened and identified six raffinose synthase (RS) genes and analyzed their physical and chemical properties, phylogenetic relationship, conserved domains, promoter cis-acting elements, and the function and genetic diversity of the gene-CDS-haplotype (gcHap). The results showed that these genes play key roles in abiotic stress response, such as OsRS5, whose expression in leaves changed significantly under high salt, drought, ABA, and MeJA treatments. In addition, the OsRS genes showed significant genetic variations in different rice populations. The main gcHaps of most OsRS loci had significant effects on key agronomic traits, and the frequency of these alleles varied significantly among different rice populations and subspecies. These findings provide direction for studying the RS gene family in other crops.
Keywords: abiotic stress; agronomic traits; genetic diversity; gene–CDS–haplotype; raffinose synthase; rice.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures







References
-
- Jing Q.K., Chen A.R., Lv Z.Y., Dong Z.H., Wang L.X., Meng X.K., Feng Y., Wan Y., Su C.Y., Cui Y.J., et al. Systematic Analysis of Galactinol Synthase and Raffinose Synthase Gene Families in Potato and Their Expression Patterns in Development and Abiotic Stress Responses. Genes. 2023;14:1344. doi: 10.3390/genes14071344. - DOI - PMC - PubMed
-
- Wang T., Wang Y.Q., Zhao J.M., Kong J.M., Zhang L.Z., Qi S.Y., Chen J.J., Chen Z.D., Zeng W., Sun W.J. Identification, Characterization and Expression Profiling of the RS Gene Family during the Withering Process of White Tea in the Tea Plant (Camellia sinensis) Reveal the Transcriptional Regulation of CsRS8. Int. J. Mol. Sci. 2023;24:202. doi: 10.3390/ijms24010202. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 2023n06020013/This research was funded by the Key Research and Development Program of Anhui Province
- 2023YFD1200900/the National Key Research and Development Program of China
- 2022AH040126/the Scientific Research Plan Major Projects of Anhui Province
- U21A20214/the National Natural Science Foundation of China
- 2021d06050002/the Improved Varieties Joint Research (Rice) Project of Anhui Province (the 14th five-year plan), and the Science and Technology Major Project of Anhui Province
LinkOut - more resources
Full Text Sources

