Identification of Tumor Suppressive miR-144-5p Targets: FAM111B Expression Accelerates the Malignant Phenotypes of Lung Adenocarcinoma
- PMID: 39337462
- PMCID: PMC11432174
- DOI: 10.3390/ijms25189974
Identification of Tumor Suppressive miR-144-5p Targets: FAM111B Expression Accelerates the Malignant Phenotypes of Lung Adenocarcinoma
Abstract
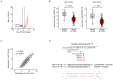
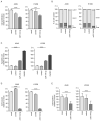
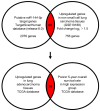
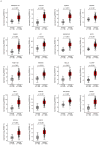
Accumulating evidence suggests that the passenger strands microRNAs (miRNAs) derived from pre-miRNAs are closely involved in cancer pathogenesis. Analysis of our miRNA expression signature of lung adenocarcinoma (LUAD) and The Cancer Genome Atlas (TCGA) data revealed that miR-144-5p (the passenger strand derived from pre-miR-144) was significantly downregulated in LUAD tissues. The aim of this study was to identify therapeutic target molecules controlled by miR-144-5p in LUAD cells. Ectopic expression assays demonstrated that miR-144-5p attenuated LUAD cell aggressiveness, e.g., inhibited cell proliferation, migration and invasion abilities, and induced cell cycle arrest and apoptotic cells. A total of 18 genes were identified as putative cancer-promoting genes controlled by miR-144-5p in LUAD cells based on our in silico analysis. We focused on a family with sequence similarity 111 member B (FAM111B) and investigated its cancer-promoting functions in LUAD cells. Luciferase reporter assay showed that expression of FAM111B was directly regulated by miR-144-5p in LUAD cells. FAM111B knockdown assays showed that LUAD cells significantly suppressed malignant phenotypes, e.g., inhibited cell proliferation, migration and invasion abilities, and induced cell cycle arrest and apoptotic cells. Furthermore, we investigated the FAM111B-mediated molecular networks in LUAD cells. Identifying target genes regulated by passenger strands of miRNAs may aid in the discovery of diagnostic markers and therapeutic targets for LUAD.
Keywords: FAM111B; lung adenocarcinoma; miR-144-5p; microRNA; passenger strand; tumor-suppressor.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

