Tracking Response and Resistance in Acute Myeloid Leukemia through Single-Cell DNA Sequencing Helps Uncover New Therapeutic Targets
- PMID: 39337490
- PMCID: PMC11432296
- DOI: 10.3390/ijms251810002
Tracking Response and Resistance in Acute Myeloid Leukemia through Single-Cell DNA Sequencing Helps Uncover New Therapeutic Targets
Abstract
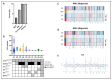
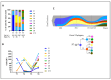
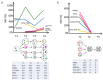
Acute myeloid leukemia (AML) is an aggressive hematologic neoplasia with a complex polyclonal architecture. Among driver lesions, those involving the FLT3 gene represent the most frequent mutations identified at diagnosis. The development of tyrosine kinase inhibitors (TKIs) has improved the clinical outcomes of FLT3-mutated patients (Pt). However, overcoming resistance to these drugs remains a challenge. To unravel the molecular mechanisms underlying therapy resistance and clonal selection, we conducted a longitudinal analysis using a single-cell DNA sequencing approach (MissionBioTapestri® platform, San Francisco, CA, USA) in two patients with FLT3-mutated AML. To this end, samples were collected at the time of diagnosis, during TKI therapy, and at relapse or complete remission. For Pt #1, disease resistance was associated with clonal expansion of minor clones, and 2nd line TKI therapy with gilteritinib provided a proliferative advantage to the clones carrying NRAS and KIT mutations, thereby responsible for relapse. In Pt #2, clonal architecture was less complex, and 1st line TKI therapy with midostaurin was able to eradicate the leukemic clones. Our results corroborate previous findings about clonal selection driven by TKIs, highlighting the importance of a deeper characterization of individual clonal architectures for choosing the best treatment plan for personalized approaches aimed at optimizing outcomes.
Keywords: FLT3 mutations; acute myeloid leukemia; clonal architecture; clonal evolution; tyrosine kinase inhibitors.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Rosnet O., Bühring H.J., Marchetto S., Rappold I., Lavagna C., Sainty D., Arnoulet C., Chabannon C., Kanz L., Hannum C., et al. Human FLT3/FLK2 receptor tyrosine kinase is expressed at the surface of normal and malignant hematopoietic cells. Leukemia. 1996;10:238–248. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

