Genetic and Functional Studies of Patients with Thyroid Dyshormonogenesis and Defects in the TSH Receptor (TSHR)
- PMID: 39337518
- PMCID: PMC11432690
- DOI: 10.3390/ijms251810032
Genetic and Functional Studies of Patients with Thyroid Dyshormonogenesis and Defects in the TSH Receptor (TSHR)
Abstract
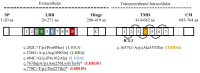
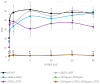
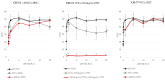
Genetic defects in the TSH receptor (TSHR) can cause poor thyroid differentiation (thyroid dysgenesis) and/or thyroid malfunction (thyroid dyshormonogenesis). The phenotype spectrum is wide: from severe congenital hypothyroidism to mild hyperthyrotropinemia. Over 250 TSHR variants have been published, many uncharacterized in vitro. We aimed to genetically characterize patients with thyroid dyshormonogenesis with TSHR defects and to study in vitro the effect of the genetic variants to establish the genotype-phenotype relationship. Pediatric patients with thyroid dyshormonogenesis (160 patients, Catalan CH neonatal screening program, confirmation TSH range: 18.4-100 mIU/L), were analyzed by a high-throughput gene panel. In vitro studies measuring the TSH-dependent cAMP-response-element activation were performed. Five patients with mild or severe thyroid dyshormonogenesis presented six TSHR variants, two unpublished. Each variant showed a different in vitro functional profile that was totally or partially deleterious. Depending on the genotype, some of the variants showed partial deficiency in both genotypes, whereas others presented a different effect. In conclusion, the percentage of patients with thyroid dyshormonogenesis and candidate variants in TSHR is 3.13%. Our in vitro studies contributed to the confirmation of the pathogenicity of the variants and highlighted the importance of studying the effect of the patient's genotype for a correct diagnostic confirmation.
Keywords: TSH receptor; TSHR; congenital hypothyroidism; functional studies; genetic variants; thyroid dyshormonogenesis; thyroid gland.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Zwaveling-Soonawala N., van Trotsenburg P. Genetics of Primary Congenital Hypothyroidism. Pediatr. Endocrinol. Rev. PER. 2018;15:200–215. - PubMed
-
- Van Trotsenburg P., Stoupa A., Léger J., Rohrer T., Peters C., Fugazzola L., Cassio A., Heinrichs C., Beauloye V., Pohlenz J., et al. Congenital Hypothyroidism: A 2020–2021 Consensus Guidelines Update—An ENDO-European Reference Network Initiative Endorsed by the European Society for Pediatric Endocrinology and the European Society for Endocrinology. Thyroid. 2021;31:387–419. doi: 10.1089/thy.2020.0333. - DOI - PMC - PubMed
-
- Szinnai G. Clinical genetics of congenital hypothyroidism. Endocr. Dev. 2014;26:60–78. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

