An 8-SNP LDL Cholesterol Polygenic Score: Associations with Cardiovascular Risk Traits, Familial Hypercholesterolemia Phenotype, and Premature Coronary Heart Disease in Central Romania
- PMID: 39337524
- PMCID: PMC11432653
- DOI: 10.3390/ijms251810038
An 8-SNP LDL Cholesterol Polygenic Score: Associations with Cardiovascular Risk Traits, Familial Hypercholesterolemia Phenotype, and Premature Coronary Heart Disease in Central Romania
Abstract
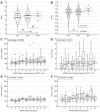
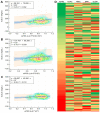
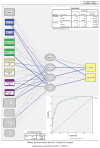
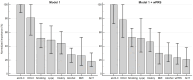
Familial hypercholesterolemia (FH) is the most significant inherited risk factor for coronary heart disease (CHD). Current guidelines focus on monogenic FH, but the polygenic form is more common and less understood. This study aimed to assess the clinical utility of an 8-SNP LDLC polygenic score in a central Romanian cohort. The cohort included 97 healthy controls and 125 patients with premature (P)CHD. The weighted LDLC polygenic risk score (wPRS) was analyzed for associations with relevant phenotypic traits, PCHD risk, and clinical FH diagnosis. The wPRS positively correlated with LDLC and DLCN scores, and LDLC concentrations could be predicted by wPRS. A trend of increasing LDLC and DLCN scores with wPRS deciles was observed. A +1 SD increase in wPRS was associated with a 36% higher likelihood of having LDLC > 190 mg/dL and increases in LDLC (+0.20 SD), DLCN score (+0.16 SD), and BMI (+0.15 SD), as well as a decrease in HDLC (-0.14 SD). Although wPRS did not predict PCHD across the entire spectrum of values, individuals above the 90th percentile were three times more likely to have PCHD compared to those within the 10th or 20th percentiles. Additionally, wPRS > 45th percentile identified "definite" clinical FH (DLCN score > 8) with 100% sensitivity and 45% specificity. The LDLC polygenic score correlates with key phenotypic traits, and individuals with high scores are more likely to have PCHD. Implementing this genetic tool may enhance risk prediction and patient stratification. These findings, the first of their kind in Romania, are consistent with the existing literature.
Keywords: LDL cholesterol; coronary heart disease; familial hypercholesterolemia; phenotype; polygenic score; single-nucleotide polymorphism.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Zarkasi K.A., Abdullah N., Murad N.A.A., Ahmad N., Jamal R. Genetic Factors for Coronary Heart Disease and Their Mechanisms: A Meta-Analysis and Comprehensive Review of Common Variants from Genome-Wide Association Studies. Diagnostics. 2022;12:2561. doi: 10.3390/diagnostics12102561. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

