Staphylococcus aureus Co-Infection in COVID-19 Patients: Virulence Genes and Their Influence on Respiratory Epithelial Cells in Light of Risk of Severe Secondary Infection
- PMID: 39337536
- PMCID: PMC11431965
- DOI: 10.3390/ijms251810050
Staphylococcus aureus Co-Infection in COVID-19 Patients: Virulence Genes and Their Influence on Respiratory Epithelial Cells in Light of Risk of Severe Secondary Infection
Abstract
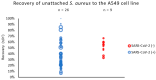
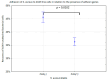
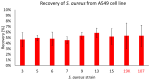
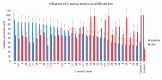
Pandemics from viral respiratory tract infections in the 20th and early 21st centuries were associated with high mortality, which was not always caused by a primary viral infection. It has been observed that severe course of infection, complications and mortality were often the result of co-infection with other pathogens, especially Staphylococcus aureus. During the COVID-19 pandemic, it was also noticed that patients infected with S. aureus had a significantly higher mortality rate (61.7%) compared to patients infected with SARS-CoV-2 alone. Our previous studies have shown that S. aureus strains isolated from patients with COVID-19 had a different protein profile than the strains in non-COVID-19 patients. Therefore, this study aims to analyze S. aureus strains isolated from COVID-19 patients in terms of their pathogenicity by analyzing their virulence genes, adhesion, cytotoxicity and penetration to the human pulmonary epithelial cell line A549. We have observed that half of the tested S. aureus strains isolated from patients with COVID-19 had a necrotizing effect on the A549 cells. The strains also showed greater variability in terms of their adhesion to the human cells than their non-COVID-19 counterparts.
Keywords: COVID-19; SARS-CoV-2; Staphylococcus aureus; adhesion.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

