HRS Facilitates Newcastle Disease Virus Replication in Tumor Cells by Promoting Viral Budding
- PMID: 39337546
- PMCID: PMC11432301
- DOI: 10.3390/ijms251810060
HRS Facilitates Newcastle Disease Virus Replication in Tumor Cells by Promoting Viral Budding
Abstract
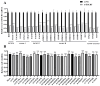
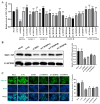
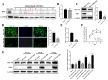
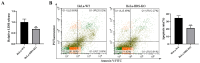
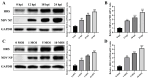
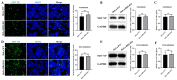
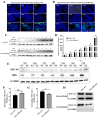
Newcastle disease virus (NDV) is a highly pathogenic avian infectious disease agent and also a promising oncolytic virus with broad application prospects. The Endosomal Sorting Complex Required for Transport (ESCRT) machinery has been increasingly recognized for its crucial role in the life cycles of enveloped viruses, influencing processes such as viral entry, replication, and budding. In this study, we employed an RNA interference screening approach to identify key ESCRT components that regulate NDV replication in tumor cells. qPCR, immunofluorescence, and Western blot assays demonstrated that knockdown of HRS, CHMP4A, CHMP4B, and CHMP4C significantly impaired NDV replication in HeLa cells, with HRS exhibiting the most pronounced inhibitory effect. Additionally, HRS knockout significantly inhibited viral budding and suppressed NDV-induced cell death in HeLa cells. Notably, NDV infection was shown to significantly upregulate HRS gene and protein expression in a time-dependent manner. In conclusion, this study systematically identifies critical ESCRT components involved in NDV replication within tumor cells, with a particular focus on the role of HRS in promoting NDV's replication by promoting viral budding, offering new insights for the development of NDV-based oncolytic therapies.
Keywords: ESCRT; HRS; NDV; viral budding; viral replication.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Chen Y., Zhu S., Liao T., Wang C., Han J., Yang Z., Lu X., Hu Z., Hu J., Wang X., et al. The HN protein of Newcastle disease virus induces cell apoptosis through the induction of lysosomal membrane permeabilization. PLoS Pathog. 2024;20:e1011981. doi: 10.1371/journal.ppat.1011981. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

