Genome-Wide Analysis and Characterization of the SDR Gene Superfamily in Cinnamomum camphora and Identification of Synthase for Eugenol Biosynthesis
- PMID: 39337570
- PMCID: PMC11432319
- DOI: 10.3390/ijms251810084
Genome-Wide Analysis and Characterization of the SDR Gene Superfamily in Cinnamomum camphora and Identification of Synthase for Eugenol Biosynthesis
Abstract
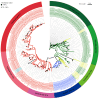
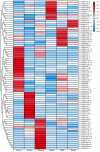
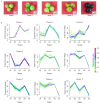
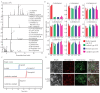
Short-chain dehydrogenase/reductases (SDRs) are the largest NAD(H)-dependent oxidoreductase superfamilies and are involved in diverse metabolisms. This study presents a comprehensive genomic analysis of the SDR superfamily in Cinnamomum camphora, a species that is one of the most significant woody essential oil plants in southern China. We identify a total of 222 CcSDR proteins and classify them into five types based on their cofactor-binding and active sites: 'atypical', 'classic', 'divergent', 'extended', and 'unknown'. Phylogenetic analysis reveals three evolutionary branches within the CcSDR proteins, and further categorization using the SDR-initiative Hidden Markov model resulted in 46 families, with the CcSDR110C, CcSDR108E, and CcSDR460A families being the most populous. Collinearity analysis identified 34 pairs of CcSDR paralogs in C. camphora, 141 pairs of SDR orthologs between C. camphora and Populus trichocarpa, and 59 pairs between C. camphora and Oryza sativa. Expression profile analysis indicates a preference for the expression of 77 CcSDR genes in specific organs such as flowers, bark, twigs, roots, leaves, or fruits. Moreover, 77 genes exhibit differential expression patterns during the four developmental stages of leaves, while 130 genes show variance across the five developmental stages of fruits. Additionally, to explore the biosynthetic mechanism of methyl eugenol, a key component of the leaf essential oil in the methyl eugenol chemotype, this study also identifies eugenol synthase (EGS) within the CcSDR460A family through an integrated strategy. Real-time quantitative PCR analysis demonstrates that the expression of CcEGS in the leaves of the methyl eugenol chemotype is more than fourfold higher compared to other chemotypes. When heterologously expressed in Escherichia coli, it catalyzes the conversion of coniferyl acetate into a mixture predominantly composed of eugenol (71.44%) and isoeugenol (21.35%). These insights pave the way for future research into the functional diversity of CcSDR genes, with a focus on secondary metabolism.
Keywords: Cinnamomum camphora; SDR460A family; eugenol synthase; expression profiles; short-chain dehydrogenase/reductase (SDR) superfamily.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Kavanagh K.L., Jörnvall H., Persson B., Oppermann U. Medium- and Short-Chain Dehydrogenase/Reductase Gene and Protein Families: The SDR Superfamily: Functional and Structural Diversity within a Family of Metabolic and Regulatory Enzymes. Cell. Mol. Life Sci. 2008;65:3895. doi: 10.1007/s00018-008-8588-y. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

