Cellular and Molecular Network Characteristics of TARM1-Related Genes in Mycobacterium tuberculosis Infections
- PMID: 39337585
- PMCID: PMC11432409
- DOI: 10.3390/ijms251810100
Cellular and Molecular Network Characteristics of TARM1-Related Genes in Mycobacterium tuberculosis Infections
Abstract
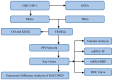
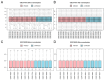
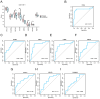
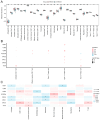
Tuberculosis (TB) is a global infectious threat, and the emergence of multidrug-resistant TB has become a major challenge in eradicating the disease that requires the discovery of new treatment strategies. This study aimed to elucidate the immune infiltration and molecular regulatory network of T cell-interacting activating receptors on myeloid cell 1 (TARM1)-related genes based on a bioinformatics analysis. The GSE114911 dataset was obtained from the Gene Expression Omnibus (GEO) and screened to identify 17 TARM1-related differentially expressed genes (TRDEGs). Genes interacting with the TRDEGs were analyzed using a Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis. A gene set enrichment analysis (GSEA) was used to identify the biological pathways significantly associated with a Mycobacterium tuberculosis (Mtb) infection. The key genes were obtained based on Cytoscape's cytoHubba plug-in. Furthermore, protein-protein interaction (PPI) networks were analyzed through STRING, while mRNA-RNA-binding protein (RBP) and mRNA-transcription factor (TF) interaction networks were developed utilizing the StarBase v3.0 and ChIPBase databases. In addition, the diagnostic significance of key genes was evaluated via receiver operating characteristic (ROC) curves, and the immune infiltration was analyzed using an ssGSEA and MCPCounter. The key genes identified in the GSE114911 dataset were confirmed in an independent GSE139825 dataset. A total of seventeen TRDEGs and eight key genes were obtained in a differential expression analysis using the cytoHubba plug-in. Through the GO and KEGG analysis, it was found that these were involved in the NF-κB, PI3K/Akt, MAPK, and other pathways related to inflammation and energy metabolism. Furthermore, the ssGSEA and MCPCounter analysis revealed a significant rise in activated T cells and T helper cells within the Mtb infection group, which were markedly associated with these key genes. This implies their potential significance in the anti-Mtb response. In summary, our results show that TRDEGs are linked to inflammation, energy metabolism, and immune cells, offering fresh insights into the mechanisms underlying TB pathogenesis and supporting further investigation into the possible molecular roles of TARM1 in TB, as well as assisting in the identification of prospective diagnostic biomarkers.
Keywords: Mycobacterium tuberculosis; TARM1; bioinformatics; biomarker; tuberculosis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
-
- Rajabalee N., Siushansian H., Weerapura M., Berton S., Berbatovci F., Hooks B., Geoffrion M., Yang D., Harper M.-E., Rayner K., et al. ATF2 orchestrates macrophage differentiation and activation to promote antibacterial responses. J. Leukoc. Biol. 2023;114:280–298. doi: 10.1093/jleuko/qiad076. - DOI - PubMed
-
- World Health Organization (WHO) Global Tuberculosis Report. World Health Organization; Geneva, Switzerland: 2023. [(accessed on 25 March 2024)]. Available online: https://www.who.int/teams/global-tuberculosis-programme/tb-reports/globa....
-
- Gandhi N.R., Moll A., Sturm A.W., Pawinski R., Govender T., Lalloo U., Zeller K., Andrews J., Friedland G. Extensively drug-resistant tuberculosis as a cause of death in patients co-infected with tuberculosis and HIV in a rural area of South Africa. Lancet. 2006;368:1575–1580. doi: 10.1016/S0140-6736(06)69573-1. - DOI - PubMed
MeSH terms
Grants and funding
- 2023Y0602/Yunnan Provincial Department of Education Science Research Fund Project
- 202401AY070001-211/Kunming Medical University, Science and Technology Department of Yunnan Province Joint project on basic research
- 82160304, 32060180, 81860644, 81560596, 31560051/National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

