Diversification of Pseudomonas aeruginosa Biofilm Populations under Repeated Phage Exposures Decreases the Efficacy of the Treatment
- PMID: 39338555
- PMCID: PMC11434582
- DOI: 10.3390/microorganisms12091880
Diversification of Pseudomonas aeruginosa Biofilm Populations under Repeated Phage Exposures Decreases the Efficacy of the Treatment
Abstract
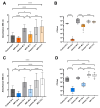
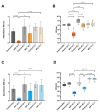
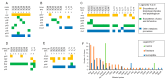
Phage therapy has been proposed as a therapeutic alternative to antibiotics for the treatment of chronic, biofilm-related P. aeruginosa infections. To gain a deeper insight into the complex biofilm-phage interactions, we investigated in the present study the effect of three successive exposures to lytic phages of biofilms formed by the reference strains PAO1 and PA14 as well as of two sequential clinical P. aeruginosa isolates from the sputum of a patient with cystic fibrosis (CF). The Calgary device was employed as a biofilm model and the efficacy of phage treatment was evaluated by measurements of the biomass stained with crystal violet (CV) and of the cell density of the biofilm bacterial population (CFU/mL) after each of the three phage exposures. The genetic alterations of P. aeruginosa isolates from biofilms exposed to phages were investigated by whole-genome sequencing. We show here that the anti-biofilm efficacy of the phage treatment decreased rapidly with repeated applications of lytic phages on P. aeruginosa strains with different genetic backgrounds. Although we observed the maintenance of a small subpopulation of sensitive cells after repeated phage treatments, a fast recruitment of mechanisms involved in the persistence of biofilms to the phage attack occurred, mainly by mutations causing alterations of the phage receptors. However, mutations causing phage-tolerant phenotypes such as alginate-hyperproducing mutants were also observed. In conclusion, a decreased anti-biofilm effect occurred after repeated exposure to lytic phages of P. aeruginosa biofilms due to the recruitment of different resistance and tolerance mechanisms.
Keywords: Pseudomonas aeruginosa; bacteriophages; biofilms; diversification of population; genomics; motility.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Hoiby N., Bjarnsholt T., Moser C., Bassi G.L., Coenye T., Donelli G., Hall-Stoodley L., Hola V., Imbert C., Kirketerp-Moller K., et al. ESCMID guideline for the diagnosis and treatment of biofilm infections 2014. Clin. Microbiol. Infect. 2015;21((Suppl. S1)):S1–S25. doi: 10.1016/j.cmi.2014.10.024. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

