Supragingival Plaque Microbiomes in a Diverse South Florida Population
- PMID: 39338595
- PMCID: PMC11434252
- DOI: 10.3390/microorganisms12091921
Supragingival Plaque Microbiomes in a Diverse South Florida Population
Abstract
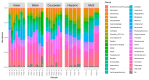
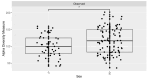
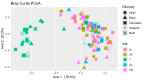
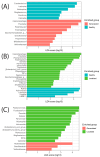
Trillions of microbes comprise the human oral cavity, collectively acting as another bodily organ. Although research is several decades into the field, there is no consensus on how oral microbiomes differ in underrepresented groups such as Hispanic, Black, and Asian populations living in the United States. Here, using 16S ribosomal RNA sequencing, we examine the bacterial ecology of supragingival plaque from four quadrants of the mouth along with a tongue swab from 26 healthy volunteers from South Florida (131 total sequences after filtering). As an area known to be a unique amalgamation of diverse cultures from across the globe, South Florida allows us to address the question of how supragingival plaque microbes differ across ethnic groups, thus potentially impacting treatment regiments related to oral issues. We assess overall phylogenetic abundance, alpha and beta diversity, and linear discriminate analysis of participants based on sex, ethnicity, sampling location in the mouth, and gingival health. Within this cohort, we find the presence of common phyla such as Firmicutes and common genera such as Streptococcus. Additionally, we find significant differences across sampling locations, sex, and gingival health. This research stresses the need for the continued incorporation of diverse populations within human oral microbiome studies.
Keywords: 16S rRNA gene sequencing; dental bacteria; human microbiota; next generation sequencing; oral health; oral microbiomes; periodontal disease; supragingival plaque.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

