Biological Evaluations and Computer-Aided Approaches of Janus Kinases 2 and 3 Inhibitors for Cancer Treatment: A Review
- PMID: 39339202
- PMCID: PMC11435443
- DOI: 10.3390/pharmaceutics16091165
Biological Evaluations and Computer-Aided Approaches of Janus Kinases 2 and 3 Inhibitors for Cancer Treatment: A Review
Abstract
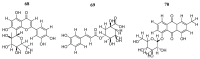
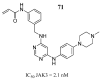
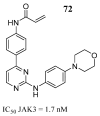
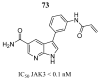
Cancer remains one of the leading diseases of mortality worldwide. Janus kinases 2/3 (JAK2/3) have been considered a drug target for the development of drugs to treat different types of cancer. JAK2/3 play a critical role in innate immunity, inflammation, and hematopoiesis by mediating the signaling of numerous cytokines, growth factors, and interferons. The current focus is to develop new selective inhibitors for each JAK type. In this review, the current strategies of computer-aided studies, and biological evaluations against JAK2/3 are addressed. We found that the new synthesized JAK2/3 inhibitors are prone to containing heterocyclic aromatic rings such as pyrimidine, pyridine, and pyrazolo [3,4-d]pyrimidine. Moreover, inhibitors of natural origin derived from plant extracts and insects have shown suitable inhibitory capacities. Computer-assisted studies have shown the important features of inhibitors for JAK2/3 binding. Biological evaluations showed that the inhibition of the JAK receptor affects its related signaling pathway. Although the reviewed compounds showed good inhibitory capacity in vitro and in vivo, more in-depth studies are needed to advance toward full approval of cancer treatments in humans.
Keywords: JAK2; JAK3; anticancer; compounds.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of the data; in the writing of the manuscript; or in the decision to publish the results.
Figures



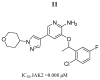

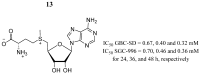

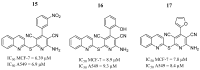

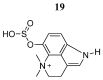
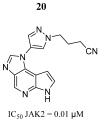

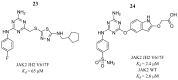

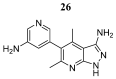
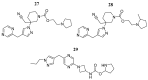
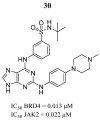
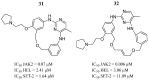

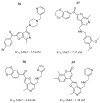
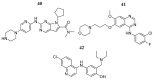


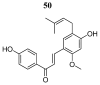
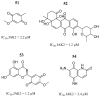
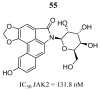


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

