Biochemical and Epigenetic Regulation of Glutamate Metabolism in Maize (Zea mays L.) Leaves under Salt Stress
- PMID: 39339624
- PMCID: PMC11434742
- DOI: 10.3390/plants13182651
Biochemical and Epigenetic Regulation of Glutamate Metabolism in Maize (Zea mays L.) Leaves under Salt Stress
Abstract
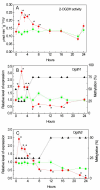
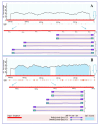
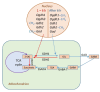
The effect of salt stress (150 mM NaCl) on the expression of genes, methylation of their promoters, and enzymatic activity of glutamate dehydrogenase (GDH), glutamate decarboxylase (GAD), and the 2-oxoglutarate (2-OG)-dehydrogenase (2-OGDH) complex was studied in maize (Zea mays L.). GDH activity increased continuously under salt stress, being 3-fold higher after 24 h. This was accompanied by the appearance of a second isoform with lower electrophoretic mobility. The expression of the Gdh1 gene strongly increased after 6-12 h of incubation, which corresponded to the demethylation of its promoter, while Gdh2 gene expression slightly increased after 2-6 h and then decreased. GAD activity gradually increased in the first 12 h, and then returned to the control level. This corresponded to the increase of Gad expression and its demethylation. Salt stress led to a 2-fold increase in the activity of 2-OGDH during the first 6 h of NaCl treatment, then the activity returned to the control level. Expression of the genes Ogdh1 and Ogdh3 peaked after 1-2 h of incubation. After 6-8 h with NaCl, the expression of these genes declined below the control levels, which correlated with the higher methylation of their promoters. We conclude that salt stress causes a redirection of the 2-OG flux to the γ-aminobutyric acid shunt via its amination to glutamate, by altering the expression of the Gdh1 and Gdh2 genes, which likely promotes the assembly of the native GDH molecule having a different subunit composition and greater affinity for 2-OG.
Keywords: 2-oxoglutarate dehydrogenase; NaCl; Zea mays L.; glutamate dehydrogenase; mitochondria; promoter methylation; salt stress; tricarboxylic acid cycle.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

