Cellular N-Myristoyl Transferases Are Required for Mammarenavirus Multiplication
- PMID: 39339839
- PMCID: PMC11436053
- DOI: 10.3390/v16091362
Cellular N-Myristoyl Transferases Are Required for Mammarenavirus Multiplication
Abstract
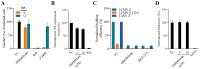
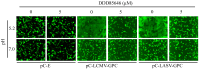
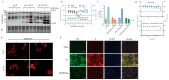
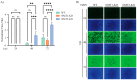
The mammarenavirus matrix Z protein plays critical roles in virus assembly and cell egress. Meanwhile, heterotrimer complexes of a stable signal peptide (SSP) together with glycoprotein subunits GP1 and GP2, generated via co-and post-translational processing of the surface glycoprotein precursor GPC, form the spikes that decorate the virion surface and mediate virus cell entry via receptor-mediated endocytosis. The Z protein and the SSP undergo N-terminal myristoylation by host cell N-myristoyltransferases (NMT1 and NMT2), and G2A mutations that prevent myristoylation of Z or SSP have been shown to affect the Z-mediated virus budding and GP2-mediated fusion activity that is required to complete the virus cell entry process. In the present work, we present evidence that the validated on-target specific pan-NMT inhibitor DDD85646 exerts a potent antiviral activity against the prototypic mammarenavirus lymphocytic choriomeningitis virus (LCMV) that correlates with reduced Z budding activity and GP2-mediated fusion activity as well as with proteasome-mediated degradation of the Z protein. The potent anti-mammarenaviral activity of DDD85646 was also observed with the hemorrhagic-fever-causing Junin (JUNV) and Lassa (LASV) mammarenaviruses. Our results support the exploration of NMT inhibition as a broad-spectrum antiviral against human pathogenic mammarenaviruses.
Keywords: LASV; LCMV; N-myristoyltransferases; antiviral; mammarenavirus; myristoylation; proteasome.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures



Update of
-
Cellular N-myristoyl transferases Are Required for Mammarenavirus Multiplication.bioRxiv [Preprint]. 2024 Aug 1:2024.08.01.606235. doi: 10.1101/2024.08.01.606235. bioRxiv. 2024. Update in: Viruses. 2024 Aug 26;16(9):1362. doi: 10.3390/v16091362. PMID: 39211253 Free PMC article. Updated. Preprint.
References
-
- Radoshitzky S.R., Buchmeier M.J., de la Torre J.C. Fields Virology: Emerging Viruses: Arenaviridae. In: Knipe D.M., Howley P.M., Whelan S., editors. Fields Virology. 7th ed. Lippincott & Wilkins; Philadelphia, PA, USA: 2022. Vol I: Emerging Viruses.
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI142985/AI/NIAID NIH HHS/United States
- R21 AI128556/AI/NIAID NIH HHS/United States
- RO1 AI142985, R21 AI128556, DoD W81XW81XWH1910496/This research was supported by NIH/NIAID grant RO1 AI142985 and R21 AI128556 to JCT, and DoD W81XW81XWH1910496 to LMS. This is manuscript 30292 from The Scripps Research Institute.
LinkOut - more resources
Full Text Sources

