Towards the Development of a Minigenome Assay for Species A Rotaviruses
- PMID: 39339871
- PMCID: PMC11437487
- DOI: 10.3390/v16091396
Towards the Development of a Minigenome Assay for Species A Rotaviruses
Abstract
RNA virus polymerases carry out multiple functions necessary for successful genome replication and transcription. A key tool for molecular studies of viral RNA-dependent RNA polymerases (RdRps) is a 'minigenome' or 'minireplicon' assay, in which viral RdRps are reconstituted in cells in the absence of full virus infection. Typically, plasmids expressing the viral polymerase protein(s) and other co-factors are co-transfected, along with a plasmid expressing an RNA encoding a fluorescent or luminescent reporter gene flanked by viral untranslated regions containing cis-acting elements required for viral RdRp recognition. This reconstitutes the viral transcription/replication machinery and allows the viral RdRp activity to be measured as a correlate of the reporter protein signal. Here, we report on the development of a 'first-generation' plasmid-based minigenome assay for species A rotavirus using a firefly luciferase reporter gene.
Keywords: RNA-dependent RNA polymerase; minigenome; reporter assay; rotavirus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
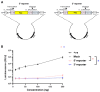
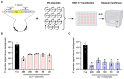


References
-
- Troeger C., Khalil I.A., Rao P.C., Cao S., Blacker B.F., Ahmed T., Armah G., Bines J.E., Brewer T.G., Colombara D.V., et al. Rotavirus Vaccination and the Global Burden of Rotavirus Diarrhea among Children Younger than 5 Years. JAMA Pediatr. 2018;172:958–965. doi: 10.1001/jamapediatrics.2018.1960. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

