Comparison of Different HIV-1 Resistance Interpretation Tools for Next-Generation Sequencing in Italy
- PMID: 39339898
- PMCID: PMC11437420
- DOI: 10.3390/v16091422
Comparison of Different HIV-1 Resistance Interpretation Tools for Next-Generation Sequencing in Italy
Abstract
Background: Next-generation sequencing (NGS) is gradually replacing Sanger sequencing for HIV genotypic drug resistance testing (GRT). This work evaluated the concordance among different NGS-GRT interpretation tools in a real-life setting.
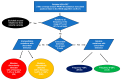
Methods: Routine NGS-GRT data were generated from viral RNA at 11 Italian laboratories with the AD4SEQ HIV-1 Solution v2 commercial kit. NGS results were interpreted by the SmartVir system provided by the kit and by two online tools (HyDRA Web and Stanford HIVdb). NGS-GRT was considered valid when the coverage was >100 reads (100×) at each PR/RT/IN resistance-associated position listed in the HIVdb 9.5.1 algorithm.
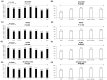
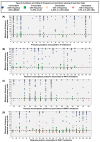
Results: Among 629 NGS-GRT, 75.2%, 74.2%, and 70.9% were valid according to SmartVir, HyDRA Web, and HIVdb. Considering at least two interpretation tools, 463 (73.6%) NGS-GRT had a valid coverage for resistance analyses. The proportion of valid samples was affected by viremia <10,000-1000 copies/mL and non-B subtypes. Mutations at an NGS frequency >10% showed fair concordance among different interpretation tools.
Conclusion: This Italian survey on NGS resistance testing suggests that viremia levels and HIV subtype affect NGS-GRT coverage. Within the current routine method for NGS-GRT, only mutations with frequency >10% seem reliably detected across different interpretation tools.
Keywords: HIV drug resistance; HIV-1 subtype; bioinformatic interpretation tools; minority variants; next-generation sequencing; viremia.
Conflict of interest statement
The authors declare no conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures

References
-
- European AIDS Clinical Society Guidelines—Version 12.0. 2023. [(accessed on 31 July 2024)]. Available online: https://www.eacsociety.org/media/guidelines-12.0.pdf.
-
- Panel on Antiretroviral Guidelines for Adults and Adolescents, Guidelines for the Use of Antiretroviral Agents in Adults and Adolescents with HIV, Department of Health and Human Services. [(accessed on 31 July 2024)];2024 Available online: https://clinicalinfo.hiv.gov/sites/default/files/guidelines/documents/ad....
-
- Parkin N.T., Avila-Rios S., Bibby D.F., Brumme C.J., Eshleman S.H., Harrigan P.R., Howison M., Hunt G., Ji H., Kantor R., et al. Multi-Laboratory Comparison of Next-Generation to Sanger-Based Sequencing for HIV-1 Drug Resistance Genotyping. Viruses. 2020;12:694. doi: 10.3390/v12070694. - DOI - PMC - PubMed
-
- Chen N.Y., Kao S.W., Liu Z.H., Wu T.S., Tsai C.L., Lin H.H., Wong W.W., Chang Y.Y., Chen S.S., Ku S.W.W. Shall I Trust the Report? Variable Performance of Sanger Sequencing Revealed by Deep Sequencing on HIV Drug Resistance Mutation Detection. Int. J. Infect. Dis. 2020;93:182–191. doi: 10.1016/j.ijid.2020.02.004. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

