SARS-CoV-2 Variants from Long-Term, Persistently Infected Immunocompromised Patients Have Altered Syncytia Formation, Temperature-Dependent Replication, and Serum Neutralizing Antibody Escape
- PMID: 39339912
- PMCID: PMC11437501
- DOI: 10.3390/v16091436
SARS-CoV-2 Variants from Long-Term, Persistently Infected Immunocompromised Patients Have Altered Syncytia Formation, Temperature-Dependent Replication, and Serum Neutralizing Antibody Escape
Abstract
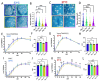
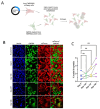
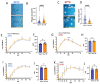
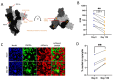
SARS-CoV-2 infection of immunocompromised individuals often leads to prolonged detection of viral RNA and infectious virus in nasal specimens, presumably due to the lack of induction of an appropriate adaptive immune response. Mutations identified in virus sequences obtained from persistently infected patients bear signatures of immune evasion and have some overlap with sequences present in variants of concern. We characterized virus isolates obtained greater than 100 days after the initial COVID-19 diagnosis from two COVID-19 patients undergoing immunosuppressive cancer therapy, wand compared them to an isolate from the start of the infection. Isolates from an individual who never mounted an antibody response specific to SARS-CoV-2 despite the administration of convalescent plasma showed slight reductions in plaque size and some showed temperature-dependent replication attenuation on human nasal epithelial cell culture compared to the virus that initiated infection. An isolate from another patient-who did mount a SARS-CoV-2 IgM response-showed temperature-dependent changes in plaque size as well as increased syncytia formation and escape from serum-neutralizing antibodies. Our results indicate that not all virus isolates from immunocompromised COVID-19 patients display clear signs of phenotypic change, but increased attention should be paid to monitoring virus evolution in this patient population.
Keywords: SARS-CoV-2; immunocompromised host; neutralizing antibody escape; syncytia; virus evolution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Clark S.A., Clark L.E., Pan J., Coscia A., McKay L.G.A., Shankar S., Johnson R.I., Brusic V., Choudhary M.C., Regan J., et al. SARS-CoV-2 Evolution in an Immunocompromised Host Reveals Shared Neutralization Escape Mechanisms. Cell. 2021;184:2605–2617.e18. doi: 10.1016/j.cell.2021.03.027. - DOI - PMC - PubMed
-
- Otto S.P., Day T., Arino J., Colijn C., Dushoff J., Li M., Mechai S., Van Domselaar G., Wu J., Earn D.J.D., et al. The Origins and Potential Future of SARS-CoV-2 Variants of Concern in the Evolving COVID-19 Pandemic. Curr. Biol. 2021;31:R918–R929. doi: 10.1016/j.cub.2021.06.049. - DOI - PMC - PubMed
-
- Braun K.M., Moreno G.K., Wagner C., Accola M.A., Rehrauer W.M., Baker D.A., Koelle K., O’Connor D.H., Bedford T., Friedrich T.C., et al. Acute SARS-CoV-2 Infections Harbor Limited within-Host Diversity and Transmit via Tight Transmission Bottlenecks. PLoS Pathog. 2021;17:e1009849. doi: 10.1371/journal.ppat.1009849. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

