N6-Methyladenosine Positively Regulates Coxsackievirus B3 Replication
- PMID: 39339923
- PMCID: PMC11437462
- DOI: 10.3390/v16091448
N6-Methyladenosine Positively Regulates Coxsackievirus B3 Replication
Abstract
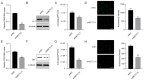
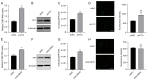
Enteroviruses such as coxsackievirus B3 are identified as a common cause of viral myocarditis, but the potential mechanism of its replication and pathogenesis are largely unknown. The genomes of a variety of viruses contain N6-methyladenosine (m6A), which plays important roles in virus replication. Here, by using the online bioinformatics tools SRAMP and indirect immunofluorescence assay (IFA), we predict that the CVB3 genome contains m6A sites and found that CVB3 infection could alter the expression and cellular localization of m6A-related proteins. Moreover, we found that 3-deazaadenosine (3-DAA), an m6A modification inhibitor, significantly decreased CVB3 replication. We also observed that the m6A methyltransferases methyltransferase-like protein 3 (METTL3) and METTL14 play positive roles in CVB3 replication, whereas m6A demethylases fat mass and obesity-associated protein (FTO) or AlkB homolog 5 (ALKBH5) have opposite effects. Knockdown of the m6A binding proteins YTH domain family protein 1 (YTHDF1), YTHDF2 and YTHDF3 strikingly decreased CVB3 replication. Finally, the m6A site mutation in the CVB3 genome decreased the replication of CVB3 compared with that in the CVB3 wild-type (WT) strain. Taken together, our results demonstrated that CVB3 could exploit m6A modification to promote viral replication, which provides new insights into the mechanism of the interaction between CVB3 and the host.
Keywords: N6-methyladenosine; coxsackievirus B3; m6A-related proteins; replication.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Peischard S., Ho H.T., Theiss C., Strutz-Seebohm N., Seebohm G. A Kidnapping Story: How Coxsackievirus B3 and Its Host Cell Interact. Cell. Physiol. Biochem. 2019;53:121–140. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

