Structural Heterogeneity of the Rabies Virus Virion
- PMID: 39339924
- PMCID: PMC11437398
- DOI: 10.3390/v16091447
Structural Heterogeneity of the Rabies Virus Virion
Abstract
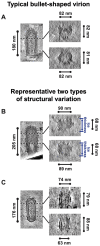
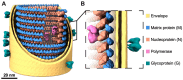
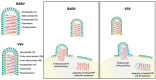
Rabies virus (RABV) is among the first recognized viruses of public health concern and has historically contributed to the development of viral vaccines. Despite these significances, the three-dimensional structure of the RABV virion remains unknown due to the challenges in isolating structurally homogenous virion samples in sufficient quantities needed for structural investigation. Here, by combining the capabilities of cryogenic electron tomography (cryoET) and microscopy (cryoEM), we determined the three-dimensional structure of the wild-type RABV virion. Tomograms of RABV virions reveal a high level of structural heterogeneity among the bullet-shaped virion particles encompassing the glycoprotein (G) trimer-decorated envelope and the nucleocapsid composed of RNA, nucleoprotein (N), and matrix protein (M). The structure of the trunk region of the virion was determined by cryoEM helical reconstruction, revealing a one-start N-RNA helix bound by a single layer of M proteins at an N:M ratio of 1. The N-M interaction differs from that in fellow rhabdovirus vesicular stomatitis virus (VSV), which features two layers of M stabilizing the N-RNA helix at an M:N ratio of 2. These differences in both M-N stoichiometry and binding allow RABV to flex its N-RNA helix more freely and point to different mechanisms of viral assembly between these two bullet-shaped rhabdoviruses.
Keywords: cryogenic electron microscopy; cryogenic electron tomography; dynamics; flexibility; rabies virus; rhabdoviruses; wild type.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Cryo EM structure of the rabies virus ribonucleoprotein complex.Sci Rep. 2019 Jul 3;9(1):9639. doi: 10.1038/s41598-019-46126-7. Sci Rep. 2019. PMID: 31270364 Free PMC article.
-
Morphogenesis of Bullet-Shaped Rabies Virus Particles Regulated by TSG101.J Virol. 2023 May 31;97(5):e0043823. doi: 10.1128/jvi.00438-23. Epub 2023 Apr 12. J Virol. 2023. PMID: 37042780 Free PMC article.
-
Three dimensional morphology of rabies virus studied by cryo-electron tomography.J Struct Biol. 2011 Oct;176(1):32-40. doi: 10.1016/j.jsb.2011.07.003. Epub 2011 Jul 19. J Struct Biol. 2011. PMID: 21784158
-
Components and Architecture of the Rhabdovirus Ribonucleoprotein Complex.Viruses. 2020 Aug 29;12(9):959. doi: 10.3390/v12090959. Viruses. 2020. PMID: 32872471 Free PMC article. Review.
-
[Rhabdoviruses].Uirusu. 2012;62(2):183-96. doi: 10.2222/jsv.62.183. Uirusu. 2012. PMID: 24153229 Review. Japanese.
References
-
- Dubos R.J. Louis Pasteur: Freelance of Science. Little, Brown and Company; Boston, MA, USA: 1950. pp. 335–336.
-
- Ng A.K., Zhang H., Tan K., Li Z., Liu J.H., Chan P.K., Li S.M., Chan W.Y., Au S.W., Joachimiak A., et al. Structure of the influenza virus A H5N1 nucleoprotein: Implications for RNA binding, oligomerization, and vaccine design. FASEB J. 2008;22:3638–3647. doi: 10.1096/fj.08-112110. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

