Mapping Transmission Dynamics and Drug Resistance Surveillance in the Cyprus HIV-1 Epidemic (2017-2021)
- PMID: 39339925
- PMCID: PMC11437465
- DOI: 10.3390/v16091449
Mapping Transmission Dynamics and Drug Resistance Surveillance in the Cyprus HIV-1 Epidemic (2017-2021)
Abstract
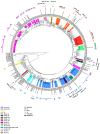
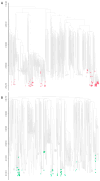
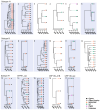
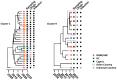
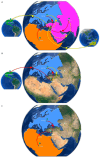
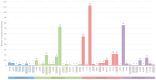
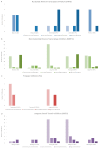
The human immunodeficiency virus type 1 (HIV-1) epidemic has been a major public health threat on a global scale since the early 1980s. Despite the introduction of combination antiretroviral therapy (cART), the incidence of new HIV-1 infections continues to rise in some regions around the world. Thus, with the continuous transmission of HIV-1 and the lack of a cure, it is imperative for molecular epidemiological studies to be performed, to monitor the infection and ultimately be able to control the spread of this virus. This work provides a comprehensive molecular epidemiological analysis of the HIV-1 infection in Cyprus, through examining 305 HIV-1 sequences collected between 9 March 2017 and 14 October 2021. Employing advanced statistical and bioinformatic techniques, the research delved deeply into understanding the transmission dynamics of the HIV-1 epidemic in Cyprus, as well as the monitoring of HIV-1's genetic diversity and the surveillance of transmitted drug resistance. The characterization of Cyprus's HIV-1 epidemic revealed a diverse landscape, comprising 21 HIV-1 group M pure subtypes and circulating recombinant forms (CRFs), alongside numerous uncharacterized recombinant strains. Subtypes A1 and B emerged as the most prevalent strains, followed by CRF02_AG. The findings of this study also revealed high levels of transmitted drug resistance (TDR) patterns, raising concerns for the efficacy of cART. The demographic profiles of individuals involved in HIV-1 transmission underscored the disproportionate burden borne by young to middle-aged Cypriot males, particularly those in the MSM community, who reported contracting the virus in Cyprus. An assessment of the spatiotemporal evolutionary dynamics illustrated the global interconnectedness of HIV-1 transmission networks, implicating five continents in the dissemination of strains within Cyprus: Europe, Africa, Asia, North America, and Oceania. Overall, this study advances the comprehension of the HIV-1 epidemic in Cyprus and highlights the importance of understanding HIV-1's transmission dynamics through continuous surveillance efforts. Furthermore, this work emphasizes the critical role of state-of-the-art bioinformatics analyses in addressing the challenges posed by HIV-1 transmission globally, laying the groundwork for public health interventions aimed at curbing its spread and improving patient outcomes.
Keywords: Cyprus; HIV-1 molecular epidemiology; HIV-1 phylodynamics; HIV-1 phylogenetics; HIV-1 phylogeographics; HIV-1 transmitted drug resistance; human immunodeficiency virus type 1 (HIV-1).
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of the data; in the writing of the manuscript; or in the decision to publish the results.
Figures


References
-
- World Health Organization (WHO) HIV: Global Situation and Trends. [(accessed on 24 January 2023)]. Available online: https://www.who.int/data/gho/data/themes/hiv-aids#:~:text=Sincethebeginn....
-
- HIV and AIDS Resources (HIV.gov) A Timeline of HIV and AIDS. [(accessed on 26 January 2023)]; Available online: https://www.hiv.gov/hiv-basics/overview/history/hiv-and-aids-timeline#ye....
-
- Joint United Nations Programme on HIV/AIDS (UNAIDS) UNAIDS Global HIV & AIDS Statistics—Fact Sheet. [(accessed on 20 January 2023)]. Available online: https://www.unaids.org/sites/default/files/media_asset/UNAIDS_FactSheet_....
-
- May M.T., Gompels M., Delpech V., Porter K., Orkin C., Kegg S., Hay P., Johnson M., Palfreeman A., Gilson R., et al. Impact on life expectancy of HIV-1 positive individuals of CD4+ cell count and viral load response to antiretroviral therapy. AIDS. 2014;28:1193–1202. doi: 10.1097/QAD.0000000000000243. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

