Next-Generation Sequencing Reveals a High Frequency of HIV-1 Minority Variants and an Expanded Drug Resistance Profile among Individuals on First-Line ART
- PMID: 39339930
- PMCID: PMC11437406
- DOI: 10.3390/v16091454
Next-Generation Sequencing Reveals a High Frequency of HIV-1 Minority Variants and an Expanded Drug Resistance Profile among Individuals on First-Line ART
Abstract
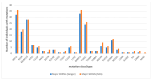
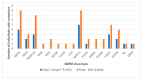
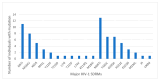
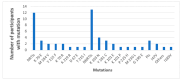
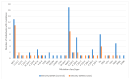
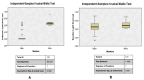
We assessed the performance and clinical relevance of Illumina MiSeq next-generation sequencing (NGS) for HIV-1 genotyping compared with Sanger sequencing (SS). We analyzed 167 participants, 45 with virologic failure (VL ≥ 1000 copies/mL), i.e., cases, and 122 time-matched participants with virologic suppression (VL < 1000 copies/mL), i.e., controls, 12 months post-ART initiation. Major surveillance drug resistance mutations (SDRMs) detected by SS were all detectable by NGS. Among cases at 12 months, SS identified SDRMs in 32/45 (71.1%) while NGS identified SDRMs among 35/45 (77.8%), increasing the number of cases with SDRMs by 3/45 (6.7%). Participants identified with, and proportions of major SDRMs increased when NGS was used. NGS vs. SS at endpoint revealed for NNRTIs: 36/45 vs. 33/45; Y181C: 26/45 vs. 24/45; K103N: 9/45 vs. 6/45 participants with SDRMs, respectively. At baseline, NGS revealed major SDRMs in 9/45 (20%) cases without SDRMs by SS. Participant MBL/043, among the nine, the following major SDRMs existed: L90M to PIs, K65R and M184V to NRTIs, and Y181C and K103N to NNRTIs. The SDRMs among the nine increased SDRMs to NRTIs, NNRTIs, and PIs. Only 43/122 (25.7%) of participants had pre-treatment minority SDRMs. Also, 24.4% of the cases vs. 26.2 of controls had minority SDRMs (p = 0.802); minority SDRMs were not associated with virologic failure. NGS agreed with SS in HIV-1 genotyping but detected additional major SDRMs and identified more participants harboring major SDRMs, expanding the HIV DRM profile of this cohort. NGS could improve HIV genotyping to guide treatment decisions for enhancing ART efficacy, a cardinal pre-requisite in the pursuit of the UNAIDS 95-95-95 targets.
Keywords: HIV-1 antiretroviral therapy; HIV-1 drug resistance; minority variants; next-generation sequencing.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures
References
-
- UNAIDS DATA 2022. Geneva: Joint United Nations Program on HIV/AIDS. 2022. [(accessed on 1 June 2024)]. Available online: https://www.unaids.org/sites/default/files/media_asset/data-book-2022_en....
-
- McMahon R. UNAIDS Issues New Fast-Track Strategy to End AIDS by 2030-EGPAF. Elizabeth Glaser Pediatric AIDS Foundation; Washington, DC, USA: 2014. [(accessed on 1 June 2024)]. Available online: https://www.pedaids.org/2014/11/20/unaids-issues-new-fast-track-strategy....
-
- WHO Surveillance of HIV Drug Resistance in Adults Receiving ART (Acquired HIV Drug Resistance) [(accessed on 5 September 2017)]. Available online: http://apps.who.int/iris/bitstream/10665/112801/1/9789241507073_eng.pdf?....
-
- Chen N.-Y., Kao S.-W., Liu Z.-H., Wu T.-S., Tsai C.-L., Lin H.-H., Wong W.-W., Chang Y.-Y., Chen S.-S., Ku S.W.-W. Shall I Trust the Report? Variable Performance of Sanger Sequencing Revealed by Deep Sequencing on HIV Drug Resistance Mutation Detection. Int. J. Infect. Dis. 2020;93:182–191. doi: 10.1016/j.ijid.2020.02.004. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

