Multiplex Microscopy Assay for Assessment of Therapeutic and Serum Antibodies against Emerging Pathogens
- PMID: 39339949
- PMCID: PMC11437451
- DOI: 10.3390/v16091473
Multiplex Microscopy Assay for Assessment of Therapeutic and Serum Antibodies against Emerging Pathogens
Abstract
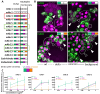
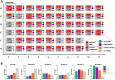
The emergence of novel pathogens, exemplified recently by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), highlights the need for rapidly deployable and adaptable diagnostic assays to assess their impact on human health and guide public health responses in future pandemics. In this study, we developed an automated multiplex microscopy assay coupled with machine learning-based analysis for antibody detection. To achieve multiplexing and simultaneous detection of multiple viral antigens, we devised a barcoding strategy utilizing a panel of HeLa-based cell lines. Each cell line expressed a distinct viral antigen, along with a fluorescent protein exhibiting a unique subcellular localization pattern for cell classification. Our robust, cell segmentation and classification algorithm, combined with automated image acquisition, ensured compatibility with a high-throughput approach. As a proof of concept, we successfully applied this approach for quantitation of immunoreactivity against different variants of SARS-CoV-2 spike and nucleocapsid proteins in sera of patients or vaccinees, as well as for the study of selective reactivity of monoclonal antibodies. Importantly, our system can be rapidly adapted to accommodate other SARS-CoV-2 variants as well as any antigen of a newly emerging pathogen, thereby representing an important resource in the context of pandemic preparedness.
Keywords: SARS-CoV-2; emerging pathogens; machine learning; monoclonal antibodies; multiplex microscopy; serology.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures



Similar articles
-
From Multiplex Serology to Serolomics-A Novel Approach to the Antibody Response against the SARS-CoV-2 Proteome.Viruses. 2021 Apr 24;13(5):749. doi: 10.3390/v13050749. Viruses. 2021. PMID: 33923338 Free PMC article.
-
SARS-CoV-2 Infection Severity Is Linked to Superior Humoral Immunity against the Spike.mBio. 2021 Jan 19;12(1):e02940-20. doi: 10.1128/mBio.02940-20. mBio. 2021. PMID: 33468695 Free PMC article.
-
Mapping SARS-CoV-2 Antibody Epitopes in COVID-19 Patients with a Multi-Coronavirus Protein Microarray.Microbiol Spectr. 2021 Oct 31;9(2):e0141621. doi: 10.1128/Spectrum.01416-21. Epub 2021 Oct 27. Microbiol Spectr. 2021. PMID: 34704808 Free PMC article.
-
Immunologic Testing for SARS-CoV-2 Infection from the Antigen Perspective.J Clin Microbiol. 2021 Apr 20;59(5):e02160-20. doi: 10.1128/JCM.02160-20. Print 2021 Apr 20. J Clin Microbiol. 2021. PMID: 33318065 Free PMC article. Review.
-
The Key to Increase Immunogenicity of Next-Generation COVID-19 Vaccines Lies in the Inclusion of the SARS-CoV-2 Nucleocapsid Protein.J Immunol Res. 2024 May 29;2024:9313267. doi: 10.1155/2024/9313267. eCollection 2024. J Immunol Res. 2024. PMID: 38939745 Free PMC article. Review.
References
-
- Ahmed F., Shafer L., Malla P., Hopkins R., Moreland S., Zviedrite N., Uzicanin A. Systematic review of empiric studies on lockdowns, workplace closures, and other non-pharmaceutical interventions in non-healthcare workplaces during the initial year of the COVID-19 pandemic: Benefits and selected unintended consequences. BMC Public Health. 2024;24:884. doi: 10.1186/s12889-024-18377-1. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

