Tuning VSV-G Expression Improves Baculovirus Integrity, Stability and Mammalian Cell Transduction Efficiency
- PMID: 39339951
- PMCID: PMC11437408
- DOI: 10.3390/v16091475
Tuning VSV-G Expression Improves Baculovirus Integrity, Stability and Mammalian Cell Transduction Efficiency
Abstract
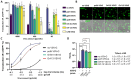
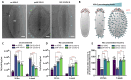
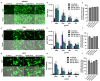
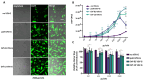
Baculoviral vectors (BVs) derived from Autographa californica multiple nucleopolyhedrovirus (AcMNPV) are an attractive tool for multigene delivery in mammalian cells, which is particularly relevant for CRISPR technologies. Most applications in mammalian cells rely on BVs that are pseudotyped with vesicular stomatitis virus G-protein (VSV-G) to promote efficient endosomal release. VSV-G expression typically occurs under the control of the hyperactive polH promoter. In this study, we demonstrate that polH-driven VSV-G expression results in BVs characterised by reduced stability, impaired morphology, and VSV-G induced toxicity at high multiplicities of transduction (MOTs) in target mammalian cells. To overcome these drawbacks, we explored five alternative viral promoters with the aim of optimising VSV-G levels displayed on the pseudotyped BVs. We report that Orf-13 and Orf-81 promoters reduce VSV-G expression to less than 5% of polH, rescuing BV morphology and stability. In a panel of human cell lines, we elucidate that BVs with reduced VSV-G support efficient gene delivery and CRISPR-mediated gene editing, at levels comparable to those obtained previously with polH VSV-G-pseudotyped BVs (polH VSV-G BV). These results demonstrate that VSV-G hyperexpression is not required for efficient transduction of mammalian cells. By contrast, reduced VSV-G expression confers similar transduction dynamics while substantially improving BV integrity, structure, and stability.
Keywords: AcMNPV; VSV-G; baculovirus; gene delivery; pseudotyping; viral vector.
Conflict of interest statement
The authors declare conflicts of interest. I.B. is shareholder in Geneva Biotech SARL, related to this correspondence. Geneva Biotech owns patents and trademarks in the field of baculovirus vector technology. All the other authors declare no conflicts of interest.
Figures

References
-
- Rohrmann G.F. The AcMNPV Genome: Gene Content, Conservation, and Function. National Center for Biotechnology Information (US); 2019. [(accessed on 27 August 2024)]. Available online: https://www.ncbi.nlm.nih.gov/books/NBK543457.
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- BB/L01386X/1/BBSRC/EPSRC Research Centre for Synthetic Biology at the University of Bristol
- BB/L014181/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- DNA-DOCK, Project No. 834631/ERC_/European Research Council/International
- BB/W013959/1/BBSRC BrisEngBio Proof-of-Concept grant
- BB/L01386X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources

