Extensive Diversity of Viruses in Millipedes Collected in the Dong Nai Biosphere Reserve (Vietnam)
- PMID: 39339962
- PMCID: PMC11437466
- DOI: 10.3390/v16091486
Extensive Diversity of Viruses in Millipedes Collected in the Dong Nai Biosphere Reserve (Vietnam)
Abstract
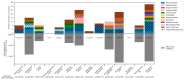
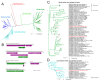
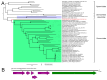
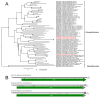
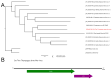
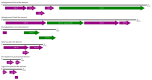
Advances in sequencing technologies and bioinformatics have led to breakthroughs in the study of virus biodiversity. Millipedes (Diplopoda, Myriapoda, Arthropoda) include more than 12,000 extant species, yet data on virus diversity in Diplopoda are scarce. This study aimed to explore the virome of the millipedes collected in the Dong Nai Biosphere Reserve in Vietnam. We studied 14 species of millipedes and managed to assemble and annotate the complete coding genomes of 16 novel viruses, the partial coding genomes of 10 more viruses, and several fragmented viral sequences, which may indicate the presence of about 54 more viruses in the studied samples. Among the complete and partial genomes, 27% were putative members of the order Picornavirales. Most of the discovered viruses were very distant from the viruses currently present in the relevant databases. At least eight viruses meet the criteria to be recognized as a new species by the International Committee on Taxonomy of Viruses, and, for two of them, a higher taxonomic status (genus and even family) can be suggested.
Keywords: Alternaviridae; Cat Tien National Park; Mitoviridae; Nairoviridae; Picornavirales; Qinviridae; Secoviridae; Xinmoviridae; tropical forest; zhaoviruses.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






Similar articles
-
Exploring Viral Diversity in a Unique South African Soil Habitat.Sci Rep. 2018 Jan 8;8(1):111. doi: 10.1038/s41598-017-18461-0. Sci Rep. 2018. PMID: 29311639 Free PMC article.
-
Massive expansion of the pig gut virome based on global metagenomic mining.NPJ Biofilms Microbiomes. 2024 Aug 29;10(1):76. doi: 10.1038/s41522-024-00554-0. NPJ Biofilms Microbiomes. 2024. PMID: 39209853 Free PMC article.
-
Virome of Bat-Infesting Arthropods: Highly Divergent Viruses in Different Vectors.J Virol. 2022 Feb 23;96(4):e0146421. doi: 10.1128/JVI.01464-21. Epub 2021 Sep 29. J Virol. 2022. PMID: 34586860 Free PMC article.
-
Current status of the Myriapod class diplopoda (millipedes): taxonomic diversity and phylogeny.Annu Rev Entomol. 2007;52:401-20. doi: 10.1146/annurev.ento.52.111805.090210. Annu Rev Entomol. 2007. PMID: 17163800 Review.
-
Virus classification - where do you draw the line?Arch Virol. 2018 Aug;163(8):2037-2046. doi: 10.1007/s00705-018-3938-z. Epub 2018 Jul 24. Arch Virol. 2018. PMID: 30039318 Free PMC article. Review.
References
Publication types
MeSH terms
Associated data
- BioProject/PRJNA1138258
- SRA/PRJNA1138258
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

