Is Autophagy a Friend or Foe in SARS-CoV-2 Infection?
- PMID: 39339967
- PMCID: PMC11437447
- DOI: 10.3390/v16091491
Is Autophagy a Friend or Foe in SARS-CoV-2 Infection?
Abstract
As obligate parasites, viruses need to hijack resources from infected cells to complete their lifecycle. The interaction between the virus and host determines the viral infection process, including viral propagation and the disease's outcome. Understanding the interaction between the virus and host factors is a basis for unraveling the intricate biological processes in the infected cells and thereby developing more efficient and targeted antivirals. Among the various fundamental virus-host interactions, autophagy plays vital and also complicated roles by directly engaging in the viral lifecycle and functioning as an anti- and/or pro-viral factor. Autophagy thus becomes a promising target against virus infection. Since the COVID-19 pandemic, there has been an accumulation of studies aiming to investigate the roles of autophagy in SARS-CoV-2 infection by using different models and from distinct angles, providing valuable information for systematically and comprehensively dissecting the interplay between autophagy and SARS-CoV-2. In this review, we summarize the advancements in the studies of the interaction between SARS-CoV-2 and autophagy, as well as detailed molecular mechanisms. We also update the current knowledge on the pharmacological strategies used to suppress SARS-CoV-2 replication through remodeling autophagy. These extensive studies on SARS-CoV-2 and autophagy can advance our understanding of virus-autophagy interaction and provide insights into developing efficient antiviral therapeutics by regulating autophagy.
Keywords: SARS-CoV-2; antivirals; autophagy; autophagy modulator; incomplete autophagy; virophagy.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
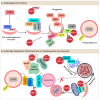
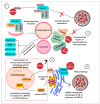
References
-
- Carabelli A.M., Peacock T.P., Thorne L.G., Harvey W.T., Hughes J., de Silva T.I., Peacock S.J., Barclay W.S., de Silva T.I., Towers G.J., et al. SARS-CoV-2 Variant Biology: Immune Escape, Transmission and Fitness. Nat. Rev. Microbiol. 2023;21:162–177. doi: 10.1038/s41579-022-00841-7. - DOI - PMC - PubMed
-
- Coronavirus Disease (COVID-19) [(accessed on 24 February 2024)]. Available online: https://www.who.int/news-room/fact-sheets/detail/coronavirus-disease-(co...
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

