Optimization of Cellular Transduction by the HIV-Based Pseudovirus Platform with Pan-Coronavirus Spike Proteins
- PMID: 39339968
- PMCID: PMC11437443
- DOI: 10.3390/v16091492
Optimization of Cellular Transduction by the HIV-Based Pseudovirus Platform with Pan-Coronavirus Spike Proteins
Abstract
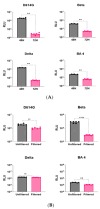
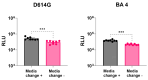
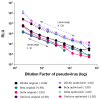
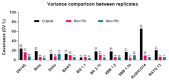
Over the past three years, new SARS-CoV-2 variants have continuously emerged, evolving to a point where an immune response against the original vaccine no longer provided optimal protection against these new strains. During this time, high-throughput neutralization assays based on pseudoviruses have become a valuable tool for assessing the efficacy of new vaccines, screening updated vaccine candidates against emerging variants, and testing the efficacy of new therapeutics such as monoclonal antibodies. Lentiviral vectors derived from HIV-1 are popular for developing pseudo and chimeric viruses due to their ease of use, stability, and long-term transgene expression. However, the HIV-based platform has lower transduction rates for pseudotyping coronavirus spike proteins than other pseudovirus platforms, necessitating more optimized methods. As the SARS-CoV-2 virus evolved, we produced over 18 variants of the spike protein for pseudotyping with an HIV-based vector, optimizing experimental parameters for their production and transduction. In this article, we present key parameters that were assessed to improve such technology, including (a) the timing and method of collection of pseudovirus supernatant; (b) the timing of host cell transduction; (c) cell culture media replenishment after pseudovirus adsorption; and (d) the centrifugation (spinoculation) parameters of the host cell+ pseudovirus mix, towards improved transduction. Additionally, we found that, for some pseudoviruses, the addition of a cationic polymer (polybrene) to the culture medium improved the transduction process. These findings were applicable across variant spike pseudoviruses that include not only SARS-CoV-2 variants, but also SARS, MERS, Alpha Coronavirus (NL-63), and bat-like coronaviruses. In summary, we present improvements in transduction efficiency, which can broaden the dynamic range of the pseudovirus titration and neutralization assays.
Keywords: coronaviruses; lentiviral pseudoviruses; neutralizing antibodies; polybrene; spinoculation; transduction efficiency.
Conflict of interest statement
Several of the authors are co-inventors of a COVID-19 recombinant protein vaccine technology held by Baylor College of Medicine (BCM). BCM has licensed this technology non-exclusively to multiple companies dedicated to improving vaccine access in low- and middle-income countries, with no patent restrictions. The authors themselves are not directly involved in the licensing negotiations led by BCM. None of the authors have additional competing financial or non-financial interests.
Figures




References
-
- D’apice L., Trovato M., Gramigna G., Colavita F., Francalancia M., Matusali G., Meschi S., Lapa D., Bettini A., Mizzoni K., et al. Comparative Analysis of the Neutralizing Activity against SARS-CoV-2 Wuhan-Hu-1 Strain and Variants of Concern: Performance Evaluation of a Pseudovirus-Based Neutralization Assay. Front. Immunol. 2022;13:981693. doi: 10.3389/fimmu.2022.981693. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

