A novel effect of sulforaphane on promoting mouse granulosa cells proliferation via the NRF2-TKT pathway
- PMID: 39341455
- PMCID: PMC12302475
- DOI: 10.1016/j.jare.2024.09.020
A novel effect of sulforaphane on promoting mouse granulosa cells proliferation via the NRF2-TKT pathway
Abstract
Introduction: Granulosa cells (GCs) is essential for maintaining follicular development. Follicle-stimulating Hormone (FSH) has been demonstrated to effectively promote GCs proliferation, driving the establishment of various superovulation techniques for animal husbandry. However, these techniques face challenges, such as high costs, hormonal imbalances, and an increased risk of early ovarian dysfunction. Therefore, it is important to investigate new methods to improve GCs proliferation.
Objectives: This study aimed to investigate the effect of sulforaphane (SFN) on ovarian GCs proliferation and the underlying mechanisms.
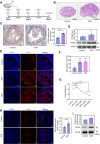
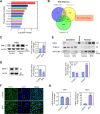
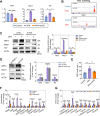
Methods: A comparative transcriptomic analysis of ovaries from the control, SFN, and FSH groups was conducted to identify the primary factors contributing to high proliferative capacity. The role of SFN in the regulation of cell proliferation has been examined in mouse ovarian GCs. Gene interference, overexpression, CUT&TAG technology, and transcriptome analyses were performed to elucidate the underlying mechanisms of the nuclear factor E2-related factor 2 (NRF2)-transketolase (TKT) axis in mediating GCs proliferation.
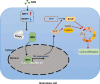
Results: Our research revealed a previously unknown function of SFN, an isothiocyanate of plant origin that is prevalent in cruciferous vegetables, in facilitating the proliferation of mouse ovarian GCs. The efficacy of SFN in enhancing GCs proliferation is similar to that of FSH. At the mechanistic level, SFN promotes NRF2 to transport to the nucleus, which subsequently activates the key enzyme of the non-oxidative pentose phosphate pathway TKT. This activation is instrumental in generating ribose 5-phosphate, a critical precursor for amino acid and nucleotide biosynthesis that underpins the proliferation of GCs.
Conclusion: Collectively, our findings delineate a novel pathway by which SFN, through the NRF2-TKT axis, enhances the nucleotide pool and thereby supports the proliferation of mouse GCs, presenting novel avenues for exploration in reproductive biology and agricultural sciences.
Keywords: Cell proliferation; Granulosa cells; NRF2–TKT; Ribose 5-phosphate; Sulforaphane.
Copyright © 2024. Published by Elsevier B.V.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
-
- Clark Z.L., Thakur M., Leach R.E., Ireland J.J. FSH dose is negatively correlated with number of oocytes retrieved: analysis of a data set with ∼650,000 ART cycles that previously identified an inverse relationship between FSH dose and live birth rate. J Assist Reprod Genet. 2021;38(7):1787–1797. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

