Plasmodium berghei HMGB1 controls the host immune responses and splenic clearance by regulating the expression of pir genes
- PMID: 39341498
- PMCID: PMC11541847
- DOI: 10.1016/j.jbc.2024.107829
Plasmodium berghei HMGB1 controls the host immune responses and splenic clearance by regulating the expression of pir genes
Abstract
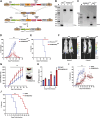
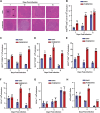
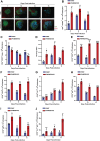
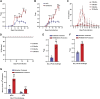
High mobility group box (HMGB) proteins belong to the high mobility group (HMG) superfamily of non-histone nuclear proteins that are involved in chromatin remodeling, regulation of gene expression, and DNA repair. When extracellular, HMGBs serve as alarmins inducing inflammation, and this is attributed to the proinflammatory activity of box B. Here, we show that Plasmodium HMGB1 has key amino acid changes in box B resulting in the loss of TNF-α stimulatory activity. Site-directed mutagenesis of the critical amino acids in box B with respect to mouse HMGB1 renders recombinant Plasmodium berghei (Pb) HMGB1 capable of inducing TNF-α release. Targeted deletion of PbHMGB1 and a detailed in vivo phenotyping show that PbHMGB1 knockout (KO) parasites can undergo asexual stage development. Interestingly, Balb/c mice-infected with PbHMGB1KO parasites display a protective phenotype with subsequent clearance of blood parasitemia and develop long-lasting protective immunity against the challenges performed with Pb wildtype parasites. The characterization of splenic responses shows prominent germinal centers leading to effective humoral responses and enhanced T follicular helper cells. There is also complete protection from experimental cerebral malaria in CBA/CaJ mice susceptible to cerebral pathogenesis with subsequent parasite clearance. Transcriptomic studies suggest the involvement of PbHMGB1 in pir expression. Our findings highlight the gene regulatory function of parasite HMGB1 and its in vivo significance in modulating the host immune responses. Further, clearance of asexual stages in PbHMGB1KO-infected mice underscores the important role of parasite HMGB1 in host immune evasion. These findings have implications in developing attenuated blood-stage vaccines for malaria.
Keywords: HMGB1; gene knockout; host immune evasion; inflammation; malaria; parasite; parasite clearance; pathogenesis; recombinant protein expression; site-directed mutagenesis.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures

References
-
- Deroost K., Pham T.T., Opdenakker G., Van den Steen P.E. The immunological balance between host and parasite in malaria. FEMS Microbiol. Rev. 2016;40:208–257. - PubMed
-
- Templeton T.J. The varieties of gene amplification, diversification and hypervariability in the human malaria parasite, Plasmodium falciparum. Mol. Biochem. Parasitol. 2009;166:109–116. - PubMed
-
- Sherman I.W., Eda S., Winograd E. Cytoadherence and sequestration in Plasmodium falciparum: defining the ties that bind. Microbes Infect. 2003;5:897–909. - PubMed
-
- Duraisingh M.T., Skillman K.M. Epigenetic variation and regulation in malaria parasites. Annu. Rev. Microbiol. 2018;72:355–375. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

