Polygenic scores stratify neurodevelopmental copy number variant carrier cognitive outcomes in the UK Biobank
- PMID: 39341812
- PMCID: PMC11438881
- DOI: 10.1038/s41525-024-00426-8
Polygenic scores stratify neurodevelopmental copy number variant carrier cognitive outcomes in the UK Biobank
Abstract
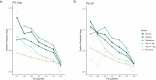
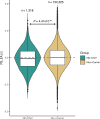
Rare copy-number variants associated with neurodevelopmental conditions (ND-CNVs) exhibit variable expressivity of clinical, physical, behavioural outcomes. Findings from clinically ascertained cohorts suggest this variability may be partly due to additional genetic variation. Here, we assessed the impact of polygenic scores (PGS) and rare variants on ND-CNV carrier fluid intelligence (FI) scores in the UK Biobank. Greater PGS for cognition (PSCog) and educational attainment (PSEA) is associated with increased FI scores in all ND-CNVs (n = 1317), 15q11.2 del. (n = 543), and 16p13.11 dup. carriers (n = 275). No association of rare variants associated with intellectual disability, autism, or putatively loss-of-function, brain-expressed genes was found. Positive predictive values in the first deciles of PScog and PSEA showed a two- to five-fold increase in the rate of low FI scores compared to baseline rates. These findings demonstrate that PGS can stratify ND-CNV carrier cognitive outcomes in a population-based cohort.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Analysis of common genetic variation and rare CNVs in the Australian Autism Biobank.Mol Autism. 2021 Feb 10;12(1):12. doi: 10.1186/s13229-020-00407-5. Mol Autism. 2021. PMID: 33568206 Free PMC article.
-
A review of the cognitive impact of neurodevelopmental and neuropsychiatric associated copy number variants.Transl Psychiatry. 2023 Apr 8;13(1):116. doi: 10.1038/s41398-023-02421-6. Transl Psychiatry. 2023. PMID: 37031194 Free PMC article. Review.
-
Cognitive performance and functional outcomes of carriers of pathogenic copy number variants: analysis of the UK Biobank.Br J Psychiatry. 2019 May;214(5):297-304. doi: 10.1192/bjp.2018.301. Epub 2019 Feb 15. Br J Psychiatry. 2019. PMID: 30767844 Free PMC article.
-
Early Manifestations of Neurodevelopmental Copy Number Variants in Children: A Population-Based Investigation.Biol Psychiatry. 2025 Mar 14:S0006-3223(25)01050-9. doi: 10.1016/j.biopsych.2025.03.004. Online ahead of print. Biol Psychiatry. 2025. PMID: 40090564
-
How does genetic variation modify ND-CNV phenotypes?Trends Genet. 2022 Feb;38(2):140-151. doi: 10.1016/j.tig.2021.07.006. Epub 2021 Aug 4. Trends Genet. 2022. PMID: 34364706 Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources

