Combinatorial optimization of the hybrid cellulase complex structure designed from modular libraries
- PMID: 39342015
- PMCID: PMC11438973
- DOI: 10.1038/s41598-024-73541-2
Combinatorial optimization of the hybrid cellulase complex structure designed from modular libraries
Abstract
Cellulase selectively recognizes cellulose surfaces and cleaves their β-1,4-glycosidic bonds. Combining hydrolysis using cellulase and fermentation can produce alternative fuels and chemical products. However, anaerobic bacteria produce only low levels of highly active cellulase complexes so-called cellulosomes. Therefore, we designed hybrid cellulase complexes from 49 biotinylated catalytic domain (CD) and 30 biotinylated cellulose-binding domain (CBD) libraries on streptavidin-conjugated nanoparticles to enhance cellulose hydrolysis by mimicking the cellulosome structure. The hybrid cellulase complex, incorporating both native CD and CBD, significantly improved reducing sugar production from cellulose compared to free native modular enzymes. The optimal CBD for each hybrid cellulase complex differed from that of the native enzyme. The most effective hybrid cellulase complex was observed with the combination of CD6-4 from Thermobifida fusca YX and CBD46 from the Bacillus halodurans C-125. The hybrid cellulase complex/CD6-4-CBD46 and -CD6-4-CBD2-5 combinations showed increased reducing sugar production. Similar results were also observed in microcrystalline cellulose degradation. Furthermore, clustering on nanoparticles enhanced enzyme thermostability. Our results demonstrate that hybrid cellulase complex structures improve enzyme function through synergistic effects and extend the lifespan of the enzyme.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




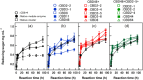
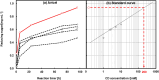
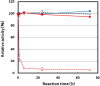
Similar articles
-
Modular Organization of the Thermobifida fusca Exoglucanase Cel6B Impacts Cellulose Hydrolysis and Designer Cellulosome Efficiency.Biotechnol J. 2017 Oct;12(10). doi: 10.1002/biot.201700205. Epub 2017 Sep 28. Biotechnol J. 2017. PMID: 28901714
-
Enhancement of cellulolytic enzyme activity by clustering cellulose binding domains on nanoscaffolds.Small. 2011 Mar 7;7(5):656-64. doi: 10.1002/smll.201002114. Epub 2011 Feb 2. Small. 2011. PMID: 21290602
-
Effect of linker length and dockerin position on conversion of a Thermobifida fusca endoglucanase to the cellulosomal mode.Appl Environ Microbiol. 2009 Dec;75(23):7335-42. doi: 10.1128/AEM.01241-09. Epub 2009 Oct 9. Appl Environ Microbiol. 2009. PMID: 19820154 Free PMC article.
-
Driving biomass breakdown through engineered cellulosomes.Bioengineered. 2015;6(4):204-8. doi: 10.1080/21655979.2015.1060379. Bioengineered. 2015. PMID: 26068180 Free PMC article. Review.
-
Cellulose, cellulases and cellulosomes.Curr Opin Struct Biol. 1998 Oct;8(5):548-57. doi: 10.1016/s0959-440x(98)80143-7. Curr Opin Struct Biol. 1998. PMID: 9818257 Review.
References
-
- Tavagnacco, L. et al. Sugar-binding sites on the surface of the carbohydrate-binding module of CBH I from Trichoderma reesei. Carbohydr. Res. 346, 839–846 (2011). - PubMed
-
- Bayer, E. A., Belaich, J. P., Shoham, Y. & Lamed, R. The cellulosomes: Multienzyme machines for degradation of plant cell wall polysaccharides. Annu. Rev. Microbiol. 58, 521–554 (2004). - PubMed
-
- Doi, R. H. & Kosugi, A. Cellulosomes: Plant-cell-wall-degrading enzyme complexes. Nat. Rev. Microbiol. 2, 541–551 (2004). - PubMed
-
- Demain, A. L., Newcomb, M. & D, H. W. Cellulase, Clostridia, and Ethanol. Thermophys. Aeromech. 22, 177–184 (2015).
-
- Fierobe, et al. Degradation of Cellulose Substrates by Cellulosome Chimeras: Substrate targeting Versus proximity of enzyme components. J. Biol. Chem. 277, 49621–49630 (2002). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

