Biomarker discovery in progressive supranuclear palsy from human cerebrospinal fluid
- PMID: 39342078
- PMCID: PMC11437921
- DOI: 10.1186/s12014-024-09507-3
Biomarker discovery in progressive supranuclear palsy from human cerebrospinal fluid
Abstract
Background: Progressive supranuclear palsy (PSP) is a neurodegenerative disorder often misdiagnosed as Parkinson's Disease (PD) due to shared symptoms. PSP is characterized by the accumulation of tau protein in specific brain regions, leading to loss of balance, gaze impairment, and dementia. Diagnosing PSP is challenging, and there is a significant demand for reliable biomarkers. Existing biomarkers, including tau protein and neurofilament light chain (NfL) levels in cerebrospinal fluid (CSF), show inconsistencies in distinguishing PSP from other neurodegenerative disorders. Therefore, the development of new biomarkers for PSP is imperative.
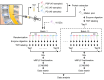
Methods: We conducted an extensive proteome analysis of CSF samples from 40 PSP patients, 40 PD patients, and 40 healthy controls (HC) using tandem mass tag-based quantification. Mass spectrometry analysis of 120 CSF samples was performed across 13 batches of 11-plex TMT experiments, with data normalization to reduce batch effects. Pathway, interactome, cell-type-specific enrichment, and bootstrap receiver operating characteristic analyses were performed to identify key candidate biomarkers.
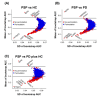
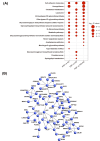
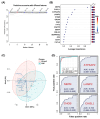
Results: We identified a total of 3,653 unique proteins. Our analysis revealed 190, 152, and 247 differentially expressed proteins in comparisons of PSP vs. HC, PSP vs. PD, and PSP vs. both PD and HC, respectively. Gene set enrichment and interactome analysis of the differentially expressed proteins in PSP CSF showed their involvement in cell adhesion, cholesterol metabolism, and glycan biosynthesis. Cell-type enrichment analysis indicated a predominance of neuronally-derived proteins among the differentially expressed proteins. The potential biomarker classification performance demonstrated that ATP6AP2 (reduced in PSP) had the highest AUC (0.922), followed by NEFM, EFEMP2, LAMP2, CHST12, FAT2, B4GALT1, LCAT, CBLN3, FSTL5, ATP6AP1, and GGH.
Conclusion: Biomarker candidate proteins ATP6AP2, NEFM, and CHI3L1 were identified as key differentiators of PSP from the other groups. This study represents the first large-scale use of mass spectrometry-based proteome analysis to identify cerebrospinal fluid (CSF) biomarkers specific to progressive supranuclear palsy (PSP) that can differentiate it from Parkinson's disease (PD) and healthy controls. Our findings lay a crucial foundation for the development and validation of reliable biomarkers, which will enhance diagnostic accuracy and facilitate early detection of PSP.
Keywords: Biomarkers; Cerebrospinal fluid; Mass spectrometry; Progressive supranuclear palsy; Proteomics.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
