Identification of functional circRNAs regulating ovarian follicle development in goats
- PMID: 39342142
- PMCID: PMC11439210
- DOI: 10.1186/s12864-024-10834-w
Identification of functional circRNAs regulating ovarian follicle development in goats
Abstract
Barkground: Circular RNAs (circRNAs) play important regulatory roles in a variety of biological processes in mammals. Multiple birth-traits in goats are affected by several factors, but the expression and function of circRNAs in follicular development of goats are not clear. In this study, we aimed to investigate the possible regulatory mechanisms of circRNA and collected five groups of large follicles (Follicle diameter > 6 mm) and small follicles (1 mm < Follicle diameter < 3 mm) from Leizhou goats in estrus for RNA sequencing.
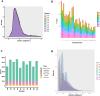
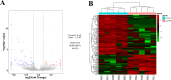
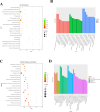
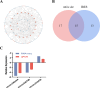
Results: RNA sequencing showed that 152 circRNAs were differentially expressed in small and large follicles. Among them, 101 circRNAs were up-regulated in large follicles and 51 circRNAs were up-regulated in small follicles. GO and KEGG enrichment analyses showed that parental genes of the differential circRNAs were significantly enriched in important pathways, such as ovarian steroidogenesis, GnRH signaling pathway, animal autophagy and oxytocin signalling pathway. BioSignal analysis revealed that 152 differentially expressed circRNAs could target 91 differential miRNAs including miR-101 family (chi-miR-101-3p, chi-miR-101-5p), miR-202 family (chi-miR-202-5p, chi-miR-202-3p),60 circRNAs with translation potential. Based on the predicted sequencing results, the ceRNA networks chicirc_008762/chi-miR-338-3p/ARHGAP18 and chicirc_040444/chi-miR-338-3p/STAR were constructed in this study. Importantly, the new gene circCFAP20DC was first discovered in goats. The EDU assay and flow cytometry results indicated that circCFAP20DC enhanced the proliferation of follicular granulosa cells(GCs). Real-time quantitative PCR and western blotting assays showed that circCFAP20DC activated the Retinoblastoma(RB) pathway and promoted the progression of granulosa cells from G1 to S phase.
Conclusion: Differential circRNAs in goat size follicles may have important biological functions for follicular development. The novel gene circCFAP20DC activates the RB pathway, promoting the progression of GCs from G1 to S phase. This, in turn, enhances the proliferation of follicular GCs in goats.
Keywords: Follicular development; Leizhou goat; RNA sequencing; Reproduction; circCFAP20DC.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Identification and Functional Analysis of circRNAs during Goat Follicular Development.Int J Mol Sci. 2024 Jul 9;25(14):7548. doi: 10.3390/ijms25147548. Int J Mol Sci. 2024. PMID: 39062792 Free PMC article.
-
Selection and Regulatory Network Analysis of Differential CircRNAs in the Hypothalamus of Goats with High and Low Reproductive Capacity.Int J Mol Sci. 2024 Sep 28;25(19):10479. doi: 10.3390/ijms251910479. Int J Mol Sci. 2024. PMID: 39408808 Free PMC article.
-
Comprehensive analysis of mRNAs and miRNAs in the ovarian follicles of uniparous and multiple goats at estrus phase.BMC Genomics. 2020 Mar 30;21(1):267. doi: 10.1186/s12864-020-6671-4. BMC Genomics. 2020. PMID: 32228439 Free PMC article.
-
Expression Profiling and Functional Analysis of Circular RNAs in Inner Mongolian Cashmere Goat Hair Follicles.Front Genet. 2021 Jun 11;12:678825. doi: 10.3389/fgene.2021.678825. eCollection 2021. Front Genet. 2021. PMID: 34178035 Free PMC article.
-
Effects of miR-101-3p on goat granulosa cells in vitro and ovarian development in vivo via STC1.J Anim Sci Biotechnol. 2020 Oct 14;11:102. doi: 10.1186/s40104-020-00506-6. eCollection 2020. J Anim Sci Biotechnol. 2020. PMID: 33072314 Free PMC article. Review.
Cited by
-
Identification and expression profile analysis of circRNAs associated with goat uterus with different fecundity during estrous cycle.BMC Genomics. 2025 Apr 7;26(1):349. doi: 10.1186/s12864-025-11489-x. BMC Genomics. 2025. PMID: 40197288 Free PMC article.
-
Missense Mutations in FDNC5 Associated with Morphometric Traits and Meat Quality in Hainan Black Goats.Animals (Basel). 2025 Feb 15;15(4):565. doi: 10.3390/ani15040565. Animals (Basel). 2025. PMID: 40003046 Free PMC article.
-
SIRT3 mediates CPT2 delactylation to enhance mitochondrial function and proliferation in goat granulosa cells.J Anim Sci Biotechnol. 2025 Jul 17;16(1):101. doi: 10.1186/s40104-025-01231-8. J Anim Sci Biotechnol. 2025. PMID: 40671144 Free PMC article.
References
-
- Angulo C, Chavez-Infante L, Reyes-Becerril M, Angulo M, Romero-Geraldo R, Llinas-Cervantes X, et al. Immunostimulatory and antioxidant effects of supplemental feeding with macroalga Sargassum spp. on goat kids. Trop anim Health pro. 2020;52(4):2023–33. - PubMed
-
- Arnal M, Robert-Granie C, Larroque H. Diversity of dairy goat lactation curves in France. J Dairy sci. 2018;101(12):11040–51. - PubMed
-
- Chen Q, Huang Y, Wang Z, Teng S, Hanif Q, Lei C, et al. Whole-genome resequencing reveals diversity and selective signals in Longlin goat. Gene. 2021;771:145371. - PubMed
-
- Liu N, Cui W, Chen M, Zhang X, Song X, Pan C. A 21-bp indel within the LLGL1 gene is significantly associated with litter size in goat. Anim Biotechnol. 2021;32(2):213–8. - PubMed
-
- Zhang Y, Cui W, Yang H, Wang M, Yan H, Zhu H, et al. A novel missense mutation (L280V) within POU1F1 gene strongly affects litter size and growth traits in goat. Theriogenology. 2019;135:198–203. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

