Structural basis for the participation of the SARS-CoV-2 nucleocapsid protein in the template switch mechanism and genomic RNA reorganization
- PMID: 39343000
- PMCID: PMC11541846
- DOI: 10.1016/j.jbc.2024.107834
Structural basis for the participation of the SARS-CoV-2 nucleocapsid protein in the template switch mechanism and genomic RNA reorganization
Abstract
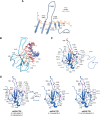
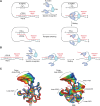
The COVID-19 pandemic has resulted in a significant toll of deaths worldwide, exceeding seven million individuals, prompting intensive research efforts aimed at elucidating the molecular mechanisms underlying the pathogenesis of SARS-CoV-2 infection. Despite the rapid development of effective vaccines and therapeutic interventions, COVID-19 remains a threat to humans due to the emergence of novel variants and largely unknown long-term consequences. Among the viral proteins, the nucleocapsid protein (N) stands out as the most conserved and abundant, playing the primary role in nucleocapsid assembly and genome packaging. The N protein is promiscuous for the recognition of RNA, yet it can perform specific functions. Here, we discuss the structural basis of specificity, which is directly linked to its regulatory role. Notably, the RNA chaperone activity of N is central to its multiple roles throughout the viral life cycle. This activity encompasses double-stranded RNA (dsRNA) annealing and melting and facilitates template switching, enabling discontinuous transcription. N also promotes the formation of membrane-less compartments through liquid-liquid phase separation, thereby facilitating the congregation of the replication and transcription complex. Considering the information available regarding the catalytic activities and binding signatures of the N protein-RNA interaction, this review focuses on the regulatory role of the SARS-CoV-2 N protein. We emphasize the participation of the N protein in discontinuous transcription, template switching, and RNA chaperone activity, including double-stranded RNA melting and annealing activities.
Keywords: RNA chaperone; RNA-binding; SARS-CoV-2; coronavirus; nucleocapsid protein.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures



Similar articles
-
The SARS-CoV-2 nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein.Nat Commun. 2021 Jan 21;12(1):502. doi: 10.1038/s41467-020-20768-y. Nat Commun. 2021. PMID: 33479198 Free PMC article.
-
Phosphoregulation of Phase Separation by the SARS-CoV-2 N Protein Suggests a Biophysical Basis for its Dual Functions.Mol Cell. 2020 Dec 17;80(6):1092-1103.e4. doi: 10.1016/j.molcel.2020.11.025. Epub 2020 Nov 20. Mol Cell. 2020. PMID: 33248025 Free PMC article.
-
N terminus of SARS-CoV-2 nonstructural protein 3 interrupts RNA-driven phase separation of N protein by displacing RNA.J Biol Chem. 2024 Nov;300(11):107828. doi: 10.1016/j.jbc.2024.107828. Epub 2024 Sep 26. J Biol Chem. 2024. PMID: 39341499 Free PMC article.
-
Unraveling the role of the nucleocapsid protein in SARS-CoV-2 pathogenesis: From viral life cycle to vaccine development.Int J Biol Macromol. 2024 Nov;279(Pt 2):135201. doi: 10.1016/j.ijbiomac.2024.135201. Epub 2024 Aug 30. Int J Biol Macromol. 2024. PMID: 39216563 Review.
-
SARS-CoV-2, the pandemic coronavirus: Molecular and structural insights.J Basic Microbiol. 2021 Mar;61(3):180-202. doi: 10.1002/jobm.202000537. Epub 2021 Jan 18. J Basic Microbiol. 2021. PMID: 33460172 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

