Local molecular and connectomic contributions of tau-related neurodegeneration
- PMID: 39343862
- PMCID: PMC11872831
- DOI: 10.1007/s11357-024-01339-1
Local molecular and connectomic contributions of tau-related neurodegeneration
Abstract
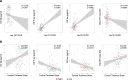
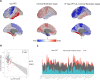
Neurodegeneration in Alzheimer's disease (AD) is known to be mostly driven by tau neurofibrillary tangles. However, both tau and neurodegeneration exhibit variability in their distribution across the brain and among individuals, and the relationship between tau and neurodegeneration might be influenced by several factors. I aimed to map local molecular and connectivity characteristics that affect the association between tau pathology and neurodegeneration. The current study was conducted on the cross-sectional tau-PET and longitudinal T1-weighted MRI scan data of 186 participants from the ADNI dataset including 71 cognitively unimpaired (CU) and 115 mild cognitive impairment (MCI) individuals. Furthermore, the normative molecular profile of a region was defined using neurotransmitter receptor densities, gene expression, T1w/T2w ratio (myelination), FDG-PET (glycolytic index, glucose metabolism, and oxygen metabolism), and synaptic density. I found that the excitatory-inhibitory (E:I) ratio, myelination, synaptic density, glycolytic index, and functional connectivity are linked with deviation in the relationship between tau and neurodegeneration. Furthermore, there was spatial similarity between tau pathology and glycolytic index, synaptic density, and functional connectivity across brain regions. The current study demonstrates that the regional susceptibility to tau-related neurodegeneration is associated with specific molecular and connectomic characteristics of the affected neural systems. I found that the molecular and connectivity architecture of the human brain is linked to the different effects of tau pathology on downstream neurodegeneration.
Keywords: Alzheimer’s disease; Connectivity; Mild cognitive impairment; Molecular; Neurodegeneration; Tau.
© 2024. The Author(s), under exclusive licence to American Aging Association.
Conflict of interest statement
Declarations. Ethical approval: The protocol for the research project has been approved by a suitably constituted Ethics Committee of an institution, and it conforms to the provisions of the Declaration of Helsinki according to the ADNI study (adni.loni.usc.edu). The STROBE checklist was followed in this observational study. Consent for publication: This manuscript has been approved for publication by all authors. Conflict of interest: The author declares no competing interests.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

