FEAtl: a comprehensive web-based expression atlas for functional genomics in tropical and subtropical fruit crops
- PMID: 39343895
- PMCID: PMC11440752
- DOI: 10.1186/s12870-024-05595-3
FEAtl: a comprehensive web-based expression atlas for functional genomics in tropical and subtropical fruit crops
Abstract
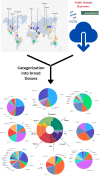
Background: Fruit crops, including tropical and subtropical fruits like Avocado (Persea americana), Fig (Ficus carica), Date Palm (Phoenix dactylifera), Mango (Mangifera indica), Guava (Psidium guajava), Papaya (Carica papaya), Pineapple (Ananas comosus), and Banana (Musa acuminata) are economically vital, contributing significantly to global agricultural output, as classified by the FAO's World Programme for the Census of Agriculture. Advancements in next-generation sequencing, have transformed fruit crop breeding by providing in-depth genomic and transcriptomic data. RNA sequencing enables high-throughput analysis of gene expression, and functional genomics, crucial for addressing horticultural challenges and enhancing fruit production. The genomic and expression data for key tropical and sub-tropical fruit crops is currently lacking a comprehensive expression atlas, revealing a significant gap in resources for horticulturists who require a unified platform with diverse datasets across various conditions and cultivars.
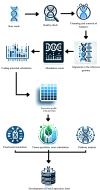
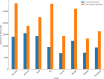
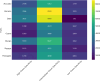
Results: The Fruit Expression Atlas (FEAtl), available at http://backlin.cabgrid.res.in/FEAtl/ , is a first-ever extensive and unified expression atlas for tropical and subtropical fruit crops developed using 3-tier architecture. The expressivity of coding and non-coding genes, encompassing 2,060 RNA-Seq samples across 91 tissue types and 177 BioProjects, it provides a comprehensive view of gene expression patterns for different tissues under various conditions. FEAtl features multiple tabs that cater to different aspects of the dataset, namely, Home, About, Analyze, Statistics, and Team and contains seven central functional modules: Transcript Information,Sample Information, Expression Profiles in FPKM and TPM, Functional Analysis, Genes Based on Tau Score, and Search for Specific Gene. The expression of a transcript of interest can be easily queried by searching by tissue ID and transcript type. Expression data can be displayed as a heat map, along with functional descriptions as well as Gene Ontology and Kyoto Encyclopedia of Genes and Genomes.
Conclusions: This atlas represents a groundbreaking compilation of a wide array of information pertaining to eight distinct fruit crops and serves as a fundamental resource for comparative analysis among different fruit species and is a catalyst for functional genomic studies. Database availability: http://backlin.cabgrid.res.in/FEAtl/ .
Keywords: Expression atlas; Fruit crops; Functional genomics; Gene expression; Genomics; Horticultural challenges; Transcriptomics.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Current status of tropical fruit breeding and genetics for three tropical fruit species cultivated in Japan: pineapple, mango, and papaya.Breed Sci. 2016 Jan;66(1):69-81. doi: 10.1270/jsbbs.66.69. Epub 2016 Jan 1. Breed Sci. 2016. PMID: 27069392 Free PMC article. Review.
-
MiSNPDb: a web-based genomic resources of tropical ecology fruit mango (Mangifera indica L.) for phylogeography and varietal differentiation.Sci Rep. 2017 Nov 2;7(1):14968. doi: 10.1038/s41598-017-14998-2. Sci Rep. 2017. PMID: 29097776 Free PMC article.
-
RNA-sequencing based gene expression landscape of guava cv. Allahabad Safeda and comparative analysis to colored cultivars.BMC Genomics. 2020 Jul 15;21(1):484. doi: 10.1186/s12864-020-06883-6. BMC Genomics. 2020. PMID: 32669108 Free PMC article.
-
De novo transcriptome assembly and annotation for gene discovery in avocado, macadamia and mango.Sci Data. 2020 Jan 8;7(1):9. doi: 10.1038/s41597-019-0350-9. Sci Data. 2020. PMID: 31913298 Free PMC article.
-
Genomic Approaches for Improvement of Tropical Fruits: Fruit Quality, Shelf Life and Nutrient Content.Genes (Basel). 2021 Nov 25;12(12):1881. doi: 10.3390/genes12121881. Genes (Basel). 2021. PMID: 34946829 Free PMC article. Review.
References
-
- Singh KM, Ahmad N, Pandey VL, Sinha DK. Impact of National Horticulture Mission on Vegetable and Fruit sectors of India. Indian J Econ Dev. 2022;66–75.
-
- Savo V, Kumbaric A, Caneva G. Grapevine (Vitis vinifera L.) Symbolism in the ancient Euro-Mediterranean cultures. Econ Bot. 2016;70(2):190–7.
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

