An automated phenotyping method for Chinese Cymbidium seedlings based on 3D point cloud
- PMID: 39343899
- PMCID: PMC11441005
- DOI: 10.1186/s13007-024-01277-1
An automated phenotyping method for Chinese Cymbidium seedlings based on 3D point cloud
Abstract
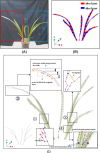
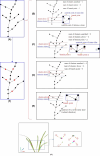
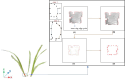
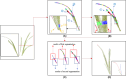
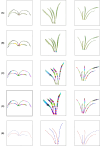
Aiming at the problems of low efficiency and high cost in determining the phenotypic parameters of Cymbidium seedlings by artificial approaches, this study proposed a fully automated measurement scheme for some phenotypic parameters based on point cloud. The key point or difficulty is to design a segmentation method for individual tillers according to the morphology-specific structure. After determining the branch points, two rounds of segmentation schemes were designed. The non-overlapping part of each tiller and the overlapping parts of each ramet are separated in the first round based on the edge point cloud-based segmentation, while in the second round, the overlapping part was sliced along the horizontal direction according to the weight ratio of the tillers above, to obtain the complete point cloud of all tillers. The core superiority of the algorithm is that the segmentation fits the tiller growth direction well, and the extracted skeleton points of tillers are close to the actual growth direction, significantly improving the prediction accuracy of the subsequent phenotypic parameters. Five phenotypic parameters, plant height, leaf number, leaf length, leaf width and leaf area, were automatically calculated. Through experiments, the accuracy of the five parameters reached 98.6%, 100%, 92.2%, 89.1%, and 82.3%, respectively, which reach the needs of various phenotypic applications.
Keywords: 3D point cloud data; Chinese Cymbidium; Phenotyping; Segmentation.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures













References
-
- Yasrab R, et al. Predicting plant growth from time-series data using deep learning. Remote Sens. 2021;13:331. - DOI
LinkOut - more resources
Full Text Sources

