UBASH3B-mediated MRPL12 Y60 dephosphorylation inhibits LUAD development by driving mitochondrial metabolism reprogramming
- PMID: 39343960
- PMCID: PMC11441236
- DOI: 10.1186/s13046-024-03181-x
UBASH3B-mediated MRPL12 Y60 dephosphorylation inhibits LUAD development by driving mitochondrial metabolism reprogramming
Abstract
Background: Metabolic reprogramming plays a pivotal role in tumorigenesis and development of lung adenocarcinoma (LUAD). However, the precise mechanisms and potential targets for metabolic reprogramming in LUAD remain elusive. Our prior investigations revealed that the mitochondrial ribosomal protein MRPL12, identified as a novel mitochondrial transcriptional regulatory gene, exerts a critical influence on mitochondrial metabolism. Despite this, the role and regulatory mechanisms underlying MRPL12's transcriptional activity in cancers remain unexplored.
Methods: Human LUAD tissues, Tp53fl/fl;KrasG12D-driven LUAD mouse models, LUAD patient-derived organoids (PDO), and LUAD cell lines were used to explored the expression and function of MRPL12. The posttranslational modification of MRPL12 was analyzed by mass spectrometry, and the oncogenic role of key phosphorylation sites of MRPL12 in LUAD development was verified in vivo and in vitro.
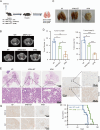
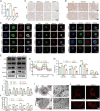
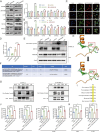
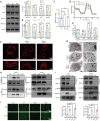
Results: MRPL12 was upregulated in human LUAD tissues, Tp53fl/fl;KrasG12D-driven LUAD tissues in mice, LUAD PDO, and LUAD cell lines, correlating with poor patient survival. Overexpression of MRPL12 significantly promoted LUAD tumorigenesis, metastasis, and PDO formation, while MRPL12 knockdown elicited the opposite phenotype. Additionally, MRPL12 deletion in a Tp53fl/fl;KrasG12D-driven mouse LUAD model conferred a notable survival advantage, delaying tumor onset and reducing malignant progression. Mechanistically, we discovered that MRPL12 promotes tumor progression by upregulating mitochondrial oxidative phosphorylation. Furthermore, we identified UBASH3B as a specific binder of MRPL12, dephosphorylating tyrosine 60 in MRPL12 (MRPL12 Y60) and inhibiting its oncogenic functions. The decrease in MRPL12 Y60 phosphorylation impeded the binding of MRPL12 to POLRMT, downregulating mitochondrial metabolism in LUAD cells. In-depth in vivo, in vitro, and organoid models validated the inhibitory effect of MRPL12 Y60 mutation on LUAD.
Conclusion: This study establishes MRPL12 as a novel oncogene in LUAD, contributing to LUAD pathogenesis by orchestrating mitochondrial metabolism reprogramming towards oxidative phosphorylation (OXPHOS). Furthermore, it confirms Y60 as a specific phosphorylation modification site regulating MRPL12's oncogenic functions, offering insights for the development of LUAD-specific targeted drugs and clinical interventions.
Keywords: LUAD; MRPL12; Metabolic reprogramming; Oxidative phosphorylation; UBASH3B.
© 2024. The Author(s).
Conflict of interest statement
Authors declare no conflict of interest.
Figures



References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209–49. - PubMed
-
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553(7689):446–54. - PubMed
-
- Howington JA, Blum MG, Chang AC, Balekian AA, Murthy SC. Treatment of stage I and II non-small cell lung cancer: Diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest. 2013;143(5 Suppl):e278S–e313S. - PubMed
-
- Calvayrac O, Pradines A, Pons E, Mazières J, Guibert N. Molecular biomarkers for lung adenocarcinoma. Eur Respir J. 2017;49(4):1601734. - PubMed
MeSH terms
Substances
Grants and funding
- 82370725/National Natural Science Foundation of China
- 82371604/National Natural Science Foundation of China
- 82271622/National Natural Science Foundation of China
- 82071569/National Natural Science Foundation of China
- 82070756/National Natural Science Foundation of China
- 82102395/National Natural Science Foundation of China
- 82200049/National Natural Science Foundation of China
- 82101639/National Natural Science Foundation of China
- ZR2021QH083/Shandong Provincial Natural Science Foundation
- ZR2023QH316/Shandong Provincial Natural Science Foundation
- ZR2021ZD35/Shandong Provincial Natural Science Foundation
- ZR2022QH349/Shandong Provincial Natural Science Foundation
- ZR2022QH374/Shandong Provincial Natural Science Foundation
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

