Cross-modal integration of bulk RNA-seq and single-cell RNA sequencing data to reveal T-cell exhaustion in colorectal cancer
- PMID: 39344205
- PMCID: PMC11439987
- DOI: 10.1111/jcmm.70101
Cross-modal integration of bulk RNA-seq and single-cell RNA sequencing data to reveal T-cell exhaustion in colorectal cancer
Abstract
Colorectal cancer (CRC) is a relatively common malignancy clinically and the second leading cause of cancer-related deaths. Recent studies have identified T-cell exhaustion as playing a crucial role in the pathogenesis of CRC. A long-standing challenge in the clinical management of CRC is to understand how T cells function during its progression and metastasis, and whether potential therapeutic targets for CRC treatment can be predicted through T cells. Here, we propose DeepTEX, a multi-omics deep learning approach that integrates cross-model data to investigate the heterogeneity of T-cell exhaustion in CRC. DeepTEX uses a domain adaptation model to align the data distributions from two different modalities and applies a cross-modal knowledge distillation model to predict the heterogeneity of T-cell exhaustion across diverse patients, identifying key functional pathways and genes. DeepTEX offers valuable insights into the application of deep learning in multi-omics, providing crucial data for exploring the stages of T-cell exhaustion associated with CRC and relevant therapeutic targets.
Keywords: T‐cell exhaustion; bulk RNA‐seq; knowledge distillation; scRNA‐seq.
© 2024 The Author(s). Journal of Cellular and Molecular Medicine published by Foundation for Cellular and Molecular Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that there are no potential conflicts of interest.
Figures




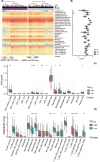

References
MeSH terms
Grants and funding
- 2024RC1062/The Science and Technology Innovation Talent Program of Hunan Province of China
- 20210002-1005 USCAT-2021-01/Research Foundation of the First Afffliated Hospital of University of South China for Advanced Talents
- 2023JJ30536/Natural Science Foundation of Hunan Province
- 2023JJ30535/Natural Science Foundation of Hunan Province
- 62171166/Data Center of Management Science, National Natural Science Foundation of China - Peking University
LinkOut - more resources
Full Text Sources
Medical

