DeepPBI-KG: a deep learning method for the prediction of phage-bacteria interactions based on key genes
- PMID: 39344712
- PMCID: PMC11440089
- DOI: 10.1093/bib/bbae484
DeepPBI-KG: a deep learning method for the prediction of phage-bacteria interactions based on key genes
Abstract
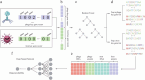
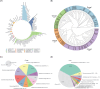
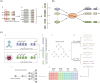
Phages, the natural predators of bacteria, were discovered more than 100 years ago. However, increasing antimicrobial resistance rates have revitalized phage research. Methods that are more time-consuming and efficient than wet-laboratory experiments are needed to help screen phages quickly for therapeutic use. Traditional computational methods usually ignore the fact that phage-bacteria interactions are achieved by key genes and proteins. Methods for intraspecific prediction are rare since almost all existing methods consider only interactions at the species and genus levels. Moreover, most strains in existing databases contain only partial genome information because whole-genome information for species is difficult to obtain. Here, we propose a new approach for interaction prediction by constructing new features from key genes and proteins via the application of K-means sampling to select high-quality negative samples for prediction. Finally, we develop DeepPBI-KG, a corresponding prediction tool based on feature selection and a deep neural network. The results show that the average area under the curve for prediction reached 0.93 for each strain, and the overall AUC and area under the precision-recall curve reached 0.89 and 0.92, respectively, on the independent test set; these values are greater than those of other existing prediction tools. The forward and reverse validation results indicate that key genes and key proteins regulate and influence the interaction, which supports the reliability of the model. In addition, intraspecific prediction experiments based on Klebsiella pneumoniae data demonstrate the potential applicability of DeepPBI-KG for intraspecific prediction. In summary, the feature engineering and interaction prediction approaches proposed in this study can effectively improve the robustness and stability of interaction prediction, can achieve high generalizability, and may provide new directions and insights for rapid phage screening for therapy.
Keywords: deep learning; machine learning; negative sample selection; phage-bacteria interaction; receptor binding protein.
© The Author(s) 2024. Published by Oxford University Press.
Figures


Similar articles
-
A Deep Learning-Based Method for Identification of Bacteriophage-Host Interaction.IEEE/ACM Trans Comput Biol Bioinform. 2021 Sep-Oct;18(5):1801-1810. doi: 10.1109/TCBB.2020.3017386. Epub 2021 Oct 7. IEEE/ACM Trans Comput Biol Bioinform. 2021. PMID: 32813660 Free PMC article.
-
DeepPL: A deep-learning-based tool for the prediction of bacteriophage lifecycle.PLoS Comput Biol. 2024 Oct 17;20(10):e1012525. doi: 10.1371/journal.pcbi.1012525. eCollection 2024 Oct. PLoS Comput Biol. 2024. PMID: 39418300 Free PMC article.
-
[Biological characteristics and genomic information of a bacteriophage against pan-drug resistant Klebsiella pneumoniae in a burn patient and its effects on bacterial biofilm].Zhonghua Shao Shang Za Zhi. 2020 Jan 20;36(1):14-23. doi: 10.3760/cma.j.issn.1009-2587.2020.01.004. Zhonghua Shao Shang Za Zhi. 2020. PMID: 32023713 Chinese.
-
Analysis of the phage sequence space: the benefit of structured information.Virology. 2007 Sep 1;365(2):241-9. doi: 10.1016/j.virol.2007.03.047. Epub 2007 May 7. Virology. 2007. PMID: 17482656 Review.
-
Global overview and major challenges of host prediction methods for uncultivated phages.Curr Opin Virol. 2021 Aug;49:117-126. doi: 10.1016/j.coviro.2021.05.003. Epub 2021 Jun 12. Curr Opin Virol. 2021. PMID: 34126465 Review.
Cited by
-
Factors Affecting Phage-Bacteria Coevolution Dynamics.Viruses. 2025 Feb 8;17(2):235. doi: 10.3390/v17020235. Viruses. 2025. PMID: 40006990 Free PMC article. Review.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

