Neuropeptide signaling network of Caenorhabditis elegans: from structure to behavior
- PMID: 39344922
- PMCID: PMC11538413
- DOI: 10.1093/genetics/iyae141
Neuropeptide signaling network of Caenorhabditis elegans: from structure to behavior
Abstract
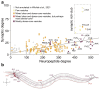
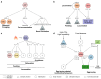
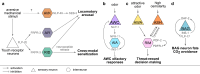
Neuropeptides are abundant signaling molecules that control neuronal activity and behavior in all animals. Owing in part to its well-defined and compact nervous system, Caenorhabditis elegans has been one of the primary model organisms used to investigate how neuropeptide signaling networks are organized and how these neurochemicals regulate behavior. We here review recent work that has expanded our understanding of the neuropeptidergic signaling network in C. elegans by mapping the evolutionary conservation, the molecular expression, the receptor-ligand interactions, and the system-wide organization of neuropeptide pathways in the C. elegans nervous system. We also describe general insights into neuropeptidergic circuit motifs and the spatiotemporal range of peptidergic transmission that have emerged from in vivo studies on neuropeptide signaling. With efforts ongoing to chart peptide signaling networks in other organisms, the C. elegans neuropeptidergic connectome can serve as a prototype to further understand the organization and the signaling dynamics of these networks at organismal level.
Keywords: Caenorhabditis elegans; behavior; neuromodulation; neuropeptide signaling network; peptide GPCRs.
© The Author(s) 2024. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest The author(s) declare no conflicts of interest.
Figures


Similar articles
-
Mapping and decoding neuropeptide signaling networks in nervous system function.Curr Opin Neurobiol. 2025 Jun;92:103027. doi: 10.1016/j.conb.2025.103027. Epub 2025 Apr 21. Curr Opin Neurobiol. 2025. PMID: 40262384 Free PMC article. Review.
-
A conserved behavioral role for a nematode interneuron neuropeptide receptor.Genetics. 2022 Jan 4;220(1):iyab198. doi: 10.1093/genetics/iyab198. Genetics. 2022. PMID: 34741504 Free PMC article.
-
The neuropeptidergic connectome of C. elegans.Neuron. 2023 Nov 15;111(22):3570-3589.e5. doi: 10.1016/j.neuron.2023.09.043. Epub 2023 Nov 6. Neuron. 2023. PMID: 37935195 Free PMC article.
-
Neuropeptidergic Signaling and Active Feeding State Inhibit Nociception in Caenorhabditis elegans.J Neurosci. 2016 Mar 16;36(11):3157-69. doi: 10.1523/JNEUROSCI.1128-15.2016. J Neurosci. 2016. PMID: 26985027 Free PMC article.
-
An overview of the insulin signaling pathway in model organisms Drosophila melanogaster and Caenorhabditis elegans.Peptides. 2021 Nov;145:170640. doi: 10.1016/j.peptides.2021.170640. Epub 2021 Aug 24. Peptides. 2021. PMID: 34450203 Review.
Cited by
-
Neuromodulation of Swarming Behavior in C. elegans: Insights into the Conserved role of Calsyntenins.bioRxiv [Preprint]. 2025 Jul 6:2025.07.06.663344. doi: 10.1101/2025.07.06.663344. bioRxiv. 2025. PMID: 40631215 Free PMC article. Preprint.
-
FLP-15 functions through the GPCR NPR-3 to regulate local and global search behaviours in Caenorhabditis elegans.bioRxiv [Preprint]. 2025 May 8:2025.05.02.651881. doi: 10.1101/2025.05.02.651881. bioRxiv. 2025. PMID: 40655019 Free PMC article. Preprint.
-
Mapping and decoding neuropeptide signaling networks in nervous system function.Curr Opin Neurobiol. 2025 Jun;92:103027. doi: 10.1016/j.conb.2025.103027. Epub 2025 Apr 21. Curr Opin Neurobiol. 2025. PMID: 40262384 Free PMC article. Review.
-
Short-term sensory memory mediates adaptation, habituation, and a paradoxical neural-behavioral transformation in C. elegans.bioRxiv [Preprint]. 2025 Aug 12:2025.08.08.669341. doi: 10.1101/2025.08.08.669341. bioRxiv. 2025. PMID: 40832180 Free PMC article. Preprint.
-
Behavioral adaptations of Caenorhabditis elegans against pathogenic threats.PeerJ. 2025 Apr 14;13:e19294. doi: 10.7717/peerj.19294. eCollection 2025. PeerJ. 2025. PMID: 40247835 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

