Mical1 deletion in tyrosinase expressing cells affects mouse running gaits
- PMID: 39344934
- PMCID: PMC11440367
- DOI: 10.1111/gbb.70004
Mical1 deletion in tyrosinase expressing cells affects mouse running gaits
Abstract
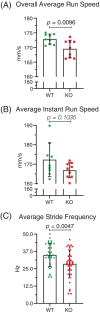
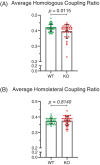
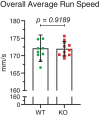
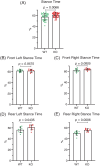
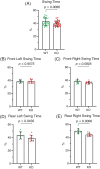
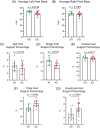
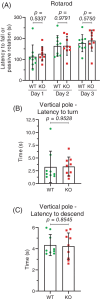
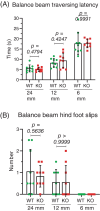
Neuronal development is a highly regulated process that is dependent on the correct coordination of cellular responses to extracellular cues. In response to semaphorin axon guidance proteins, the MICAL1 protein is stimulated to produce reactive oxygen species that oxidize actin on specific methionine residues, leading to filamentous actin depolymerization and consequent changes in neuronal growth cone dynamics. Crossing genetically modified mice homozygous for floxed Mical1 (Mical1fl/fl) alleles with transgenic mice expressing Cre recombinase under the control of a tyrosinase gene enhancer/promoter (Tyr::Cre) enabled conditional Mical1 deletion. Immunohistochemical analysis showed Mical1 expression in the cerebellum, which plays a prominent role in the coordination of motor movements, with reduced Mical1 expression in Mical1fl/fl mice co-expressing Tyr::Cre. Analysis of the gaits of mice running on a treadmill showed that both male and female Mical1fl/fl, Tyr::Cre mutant mice had significant alterations to their striding patterns relative to wild-type mice, although the specific aspects of their altered gaits differed between the sexes. Additional motor tests that involved movement on a rotating rod, descending a vertical pole, or crossing a balance beam did not show significant differences between the genotypes, suggesting that the effect of the Mical1fl/fl, Tyr::Cre genetic modifications was only manifested during specific highly coordinated movements that contribute to running. These findings indicate that there is a behavioral consequence in Mical1fl/fl, Tyr::Cre mutant mice that affects motor control as manifested by alterations in their gait.
Keywords: Mical1; actin; cytoskeleton; gait; genetic modification; reactive oxygen species; running; stride; tyrosinase.
© 2024 The Author(s). Genes, Brain and Behavior published by International Behavioural and Neural Genetics Society and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Cheever TR, Ervasti JM. Actin isoforms in neuronal development and function. Int Rev Cell Mol Biol. 2013;301:157‐213. - PubMed
-
- Goellner B, Aberle H. The synaptic cytoskeleton in development and disease. Dev Neurobiol. 2012;72(1):111‐125. - PubMed
-
- Schneider F, Metz I, Rust MB. Regulation of actin filament assembly and disassembly in growth cone motility and axon guidance. Brain Res Bull. 2023;192:21‐35. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

