Analytical Methods to Evaluate RNA Circularization Efficiency
- PMID: 39345227
- PMCID: PMC11662197
- DOI: 10.1002/elps.202400067
Analytical Methods to Evaluate RNA Circularization Efficiency
Abstract
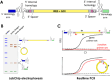
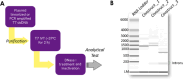
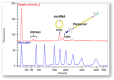
Circular RNAs (circRNAs) have emerged as pivotal players in RNA therapeutics. Unlike linear counterparts, circRNAs possess a closed-loop structure, conferring them with enhanced stability and resistance to degradation. Ribozyme-based strategy stands out as the predominant method for synthetic circRNA production, by precisely cleaving and promoting the formation of a covalent circular structure. However, there is still a lack of analytical methods that can provide high-throughput and quantitative analysis to facilitate the circRNA vector engineering process. In the report, we detail analytical methods to characterize and evaluate ribozyme-based RNA circularization efficiency. Our approach will capture the attention of researchers interested in optimizing RNA circularization efficiency, as well as those focused on exploring key elements for ribozyme catalytic activity.
Keywords: circular RNA; linear RNA; microfluidic capillary electrophoresis; vector engineering.
© 2024 Revvity, Inc. ELECTROPHORESIS published by Wiley‐VCH GmbH.
Conflict of interest statement
All authors listed are employees of Revvity, Inc.
Figures

Similar articles
-
In Vitro Self-Circularization Methods Based on Self-Splicing Ribozyme.Int J Mol Sci. 2024 Aug 30;25(17):9437. doi: 10.3390/ijms25179437. Int J Mol Sci. 2024. PMID: 39273386 Free PMC article. Review.
-
Efficient circularization of protein-encoding RNAs via a novel cis-splicing system.Nucleic Acids Res. 2024 Sep 23;52(17):10400-10415. doi: 10.1093/nar/gkae711. Nucleic Acids Res. 2024. PMID: 39162233 Free PMC article.
-
Developing an enhanced chimeric permuted intron-exon system for circular RNA therapeutics.Theranostics. 2024 Sep 9;14(15):5869-5882. doi: 10.7150/thno.98214. eCollection 2024. Theranostics. 2024. PMID: 39346546 Free PMC article.
-
Non-enzymatic in vitro production of circular hammerhead ribozyme targeting the template region of human telomerase RNA.Nucleic Acids Symp Ser (Oxf). 2009;(53):275-6. doi: 10.1093/nass/nrp138. Nucleic Acids Symp Ser (Oxf). 2009. PMID: 19749367
-
Thrown for a (stem) loop: How RNA structure impacts circular RNA regulation and function.Methods. 2021 Dec;196:56-67. doi: 10.1016/j.ymeth.2021.02.019. Epub 2021 Mar 2. Methods. 2021. PMID: 33662561 Review.
References
-
- Rohner E., Yang R., Foo K. S., Goedel A., and Chien K. R., “Unlocking the Promise of mRNA Therapeutics,” Nature Biotechnology 40 (2022): 1586–1600. - PubMed
-
- Barbier A. J., Jiang A. Y., Zhang P., Wooster R., and Anderson D. G., “The Clinical Progress of mRNA Vaccines and Immunotherapies,” Nature Biotechnology 40 (2022): 840–854. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

