This is a preprint.
Cell differentiation controls iron assimilation in a choanoflagellate
- PMID: 39345370
- PMCID: PMC11429873
- DOI: 10.1101/2024.05.25.595918
Cell differentiation controls iron assimilation in a choanoflagellate
Update in
-
Cell differentiation controls iron assimilation in the choanoflagellate Salpingoeca rosetta.mSphere. 2025 Mar 25;10(3):e0091724. doi: 10.1128/msphere.00917-24. Epub 2025 Feb 26. mSphere. 2025. PMID: 40008892 Free PMC article.
Abstract
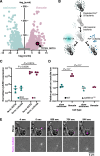
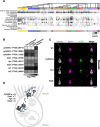
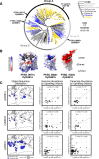
Marine microeukaryotes have evolved diverse cellular features that link their life histories to surrounding environments. How those dynamic life histories intersect with the ecological functions of microeukaryotes remains a frontier to understand their roles in essential biogeochemical cycles1,2. Choanoflagellates, phagotrophs that cycle nutrients through filter feeding, provide models to explore this intersection, for many choanoflagellate species transition between life history stages by differentiating into distinct cell types3-6. Here we report that cell differentiation in the marine choanoflagellate Salpingoeca rosetta endows one of its cell types with the ability to utilize insoluble ferric colloids for improved growth through the expression of a cytochrome b561 iron reductase (cytb561a). This gene is an ortholog of the mammalian duodenal cytochrome b561 (DCYTB) that reduces ferric cations prior to their uptake in gut epithelia7 and is part of an iron utilization toolkit that choanoflagellates and their closest living relatives, the animals, inherited from a last common eukaryotic ancestor. In a database of oceanic metagenomes8,9, the abundance of cytb561a transcripts from choanoflagellates positively correlates with upwellings, which are a major source of ferric colloids in marine environments10. As this predominant form of iron11,12 is largely inaccessible to cell-walled microbes13,14, choanoflagellates and other phagotrophic eukaryotes may serve critical ecological roles by first acquiring ferric colloids through phagocytosis and then cycling this essential nutrient through iron utilization pathways13-15. These findings provide insight into the ecological roles choanoflagellates perform and inform reconstructions of early animal evolution where functionally distinct cell types became an integrated whole at the origin of animal multicellularity16-22.
Figures

References
-
- Worden A. Z. et al. Rethinking the marine carbon cycle: Factoring in the multifarious lifestyles of microbes. Science 347, (2015). - PubMed
-
- Leadbeater B. S. C. Observations on the life-history and ultrastructure of the marine choanoflagellate Choanoeca perplexa Ellis. J. Mar. Biol. Assoc. U. K. 57, 285–301 (1977).
-
- Leadbeater B. S. C. The Choanoflagellates: Evolution, Biology, and Ecology. (Cambridge University Press, Cambridge, United Kingdom, 2015).
-
- Reyes-Rivera J. et al. Nitric oxide signaling controls collective contractions in a colonial choanoflagellate. Curr. Biol. 32, 2539–2547.e5 (2022). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
