This is a preprint.
The mechanism of peptidoglycan O-acetylation in Gram-negative bacteria typifies bacterial MBOAT-SGNH acyltransferases
- PMID: 39345430
- PMCID: PMC11429678
- DOI: 10.1101/2024.09.17.613324
The mechanism of peptidoglycan O-acetylation in Gram-negative bacteria typifies bacterial MBOAT-SGNH acyltransferases
Update in
-
The mechanism of peptidoglycan O-acetylation in Gram-negative bacteria typifies bacterial MBOAT-SGNH acyltransferases.J Biol Chem. 2025 Jun;301(6):108531. doi: 10.1016/j.jbc.2025.108531. Epub 2025 Apr 23. J Biol Chem. 2025. PMID: 40280421 Free PMC article.
Abstract
Bacterial cell envelope polymers are commonly modified with acyl groups that provide fitness advantages. Many polymer acylation pathways involve pairs of membrane-bound O-acyltransferase (MBOAT) and SGNH family proteins. As an example, the MBOAT protein PatA and the SGNH protein PatB are required in Gram-negative bacteria for peptidoglycan O-acetylation. The mechanism for how MBOAT-SGNH transferases move acyl groups from acyl-CoA donors made in the cytoplasm to extracellular polymers is unclear. Using the peptidoglycan O-acetyltransferase proteins PatAB, we explore the mechanism of MBOAT-SGNH pairs. We find that the MBOAT protein PatA catalyzes auto-acetylation of an invariant Tyr residue in its conserved C-terminal hexapeptide motif. We also show that PatB can use a synthetic hexapeptide containing an acetylated tyrosine to donate an acetyl group to a peptidoglycan mimetic. Finally, we report the structure of PatB, finding that it has structural features that shape its activity as an O-acetyltransferase and distinguish it from other SGNH esterases and hydrolases. Taken together, our results support a model for peptidoglycan acylation in which a tyrosine-containing peptide at the MBOAT's C-terminus shuttles an acyl group from the MBOAT active site to the SGNH active site, where it is transferred to peptidoglycan. This model likely applies to other systems containing MBOAT-SGNH pairs, such as those that O-acetylate alginate, cellulose, and secondary cell wall polysaccharides. The use of an acyl-tyrosine intermediate for MBOAT-SGNH acyl transfer is also shared with AT3-SGNH proteins, a second major group of acyltransferases that modify cell envelope polymers.
Keywords: Bacterial cell wall; O-acetylation; O-acetyltransferase; X-ray crystallography; peptidoglycan.
Conflict of interest statement
Competing Interest Statement: The authors declare no competing interest.
Figures

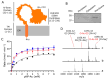
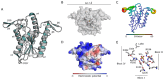
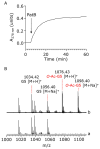

References
-
- Vollmer W., Blanot D. & De Pedro M. A. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32, 149–167 (2008). - PubMed
-
- Whitfield C. & Trent M. . S. Biosynthesis and export of bacterial lipopolysaccharides. Annu. Rev. Biochem. 83, 99–128 (2014). - PubMed
-
- Bera A., Herbert S., Jakob A., Vollmer W. & Götz F. Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol. Microbiol. 55, 778–787 (2005). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
