This is a preprint.
Quantitative analysis of non-histone lysine methylation sites and lysine demethylases in breast cancer cell lines
- PMID: 39345446
- PMCID: PMC11429713
- DOI: 10.1101/2024.09.18.613658
Quantitative analysis of non-histone lysine methylation sites and lysine demethylases in breast cancer cell lines
Update in
-
Quantitative Analysis of Nonhistone Lysine Methylation Sites and Lysine Demethylases in Breast Cancer Cell Lines.J Proteome Res. 2025 Feb 7;24(2):550-561. doi: 10.1021/acs.jproteome.4c00685. Epub 2025 Jan 8. J Proteome Res. 2025. PMID: 39778878
Abstract
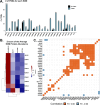
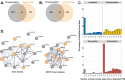
Growing evidence shows that lysine methylation is a widespread protein post-translational modification that regulates protein function on histone and non-histone proteins. Numerous studies have demonstrated that dysregulation of lysine methylation mediators contributes to cancer growth and chemotherapeutic resistance. While changes in histone methylation are well documented with extensive analytical techniques available, there is a lack of high-throughput methods to reproducibly quantify changes in the abundances of the mediators of lysine methylation and non-histone lysine methylation (Kme) simultaneously across multiple samples. Recent studies by our group and others have demonstrated that antibody enrichment is not required to detect lysine methylation, prompting us to investigate the use of Tandem Mass Tag (TMT) labeling for global Kme quantification sans antibody enrichment in four different breast cancer cell lines (MCF-7, MDA-MB-231, HCC1806, and MCF10A). To improve the quantification of KDMs, we incorporated a lysine demethylase (KDM) isobaric trigger channel, which enabled 96% of all KDMs to be quantified while simultaneously quantifying 326 Kme sites. Overall, 142 differentially abundant Kme sites and eight differentially abundant KDMs were identified between the four cell lines, revealing cell line-specific patterning.
Keywords: TMT; lysine demethylases; lysine methylation.
Figures



References
-
- Liu S.; Hausmann S.; Carlson S. M.; Fuentes M. E.; Francis J. W.; Pillai R.; Lofgren S. M.; Hulea L.; Tandoc K.; Lu J.; Li A.; Nguyen N. D.; Caporicci M.; Kim M. P.; Maitra A.; Wang H.; Wistuba I. I.; Porco J. A.; Bassik M. C.; Elias J. E.; Song J.; Topisirovic I.; Van Rechem C.; Mazur P. K.; Gozani O. METTL13 Methylation of eEF1A Increases Translational Output to Promote Tumorigenesis. Cell 2019, 176 (3), 491–504.e21. 10.1016/j.cell.2018.11.038. - DOI - PMC - PubMed
-
- Park S. H.; Fong K.; Kim J.; Wang F.; Lu X.; Lee Y.; Brea L. T.; Wadosky K.; Guo C.; Abdulkadir S. A.; Crispino J. D.; Fang D.; Ntziachristos P.; Liu X.; Li X.; Wan Y.; Goodrich D. W.; Zhao J. C.; Yu J. Posttranslational Regulation of FOXA1 by Polycomb and BUB3/USP7 Deubiquitin Complex in Prostate Cancer. Sci. Adv. 2021, 7 (15), eabe2261. 10.1126/sciadv.abe2261. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
