This is a preprint.
Endogenous hydrogen peroxide positively regulates secretion of a gut-derived peptide in neuroendocrine potentiation of the oxidative stress response in C. elegans
- PMID: 39345448
- PMCID: PMC11429608
- DOI: 10.1101/2024.04.03.587937
Endogenous hydrogen peroxide positively regulates secretion of a gut-derived peptide in neuroendocrine potentiation of the oxidative stress response in C. elegans
Update in
-
Endogenous hydrogen peroxide positively regulates secretion of a gut-derived peptide in neuroendocrine potentiation of the oxidative stress response in Caenorhabditis elegans.Elife. 2024 Dec 5;13:RP97503. doi: 10.7554/eLife.97503. Elife. 2024. PMID: 39636673 Free PMC article.
Abstract
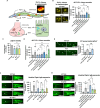
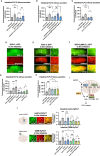
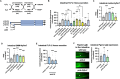
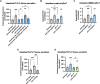
The gut-brain axis mediates bidirectional signaling between the intestine and the nervous system and is critical for organism-wide homeostasis. Here we report the identification of a peptidergic endocrine circuit in which bidirectional signaling between neurons and the intestine potentiates the activation of the antioxidant response in C. elegans in the intestine. We identify a FMRF-amide-like peptide, FLP-2, whose release from the intestine is necessary and sufficient to activate the intestinal oxidative stress response by promoting the release of the antioxidant FLP-1 neuropeptide from neurons. FLP-2 secretion from the intestine is positively regulated by endogenous hydrogen peroxide (H2O2) produced in the mitochondrial matrix by sod-3/superoxide dismutase, and is negatively regulated by prdx-2/peroxiredoxin, which depletes H2O2 in both the mitochondria and cytosol. H2O2 promotes FLP-2 secretion through the DAG and calciumdependent protein kinase C family member pkc-2 and by the SNAP25 family member aex-4 in the intestine. Together, our data demonstrate a role for intestinal H2O2 in promoting inter-tissue antioxidant signaling through regulated neuropeptide-like protein exocytosis in a gut-brain axis to activate the oxidative stress response.
Keywords: AEX-4; C. elegans; PKC-2; PRDX-2; SNAP25; SOD-1; SOD-3; TRX-3; hydrogen peroxide; mitochondria; neuropeptide; oxidative stress; peroxiredoxin; protein kinase C; superoxide dismutase; thioredoxin.
Figures


Similar articles
-
Endogenous hydrogen peroxide positively regulates secretion of a gut-derived peptide in neuroendocrine potentiation of the oxidative stress response in Caenorhabditis elegans.Elife. 2024 Dec 5;13:RP97503. doi: 10.7554/eLife.97503. Elife. 2024. PMID: 39636673 Free PMC article.
-
Sphingosine Kinase Regulates Neuropeptide Secretion During the Oxidative Stress-Response Through Intertissue Signaling.J Neurosci. 2018 Sep 19;38(38):8160-8176. doi: 10.1523/JNEUROSCI.0536-18.2018. Epub 2018 Aug 6. J Neurosci. 2018. PMID: 30082417 Free PMC article.
-
Mitochondrial hydrogen peroxide positively regulates neuropeptide secretion during diet-induced activation of the oxidative stress response.Nat Commun. 2021 Apr 16;12(1):2304. doi: 10.1038/s41467-021-22561-x. Nat Commun. 2021. PMID: 33863916 Free PMC article.
-
Riding the tiger - physiological and pathological effects of superoxide and hydrogen peroxide generated in the mitochondrial matrix.Crit Rev Biochem Mol Biol. 2020 Dec;55(6):592-661. doi: 10.1080/10409238.2020.1828258. Epub 2020 Nov 4. Crit Rev Biochem Mol Biol. 2020. PMID: 33148057 Review.
-
A radical shift in perspective: mitochondria as regulators of reactive oxygen species.J Exp Biol. 2017 Apr 1;220(Pt 7):1170-1180. doi: 10.1242/jeb.132142. J Exp Biol. 2017. PMID: 28356365 Review.
References
-
- Ahier Arnaud, Dai Chuan-Yang, Tweedie Andrea, Bezawork-Geleta Ayenachew, Kirmes Ina, and Zuryn Steven. 2018. “Affinity Purification of Cell-Specific Mitochondria from Whole Animals Resolves Patterns of Genetic Mosaicism.” Nature Cell Biology 20(3):352-. doi: 10.1038/s41556-017-0023-x. - DOI - PubMed
-
- Ailion Michael, Hannemann Mandy, Dalton Susan, Pappas Andrea, Watanabe Shigeki, Hegermann Jan, Liu Qiang, Han Hsiao-Fen, Gu Mingyu, Goulding Morgan Q., Sasidharan Nikhil, Schuske Kim, Hullett Patrick, Eimer Stefan, and Jorgensen Erik M.. 2014. “Two Rab2 Interactors Regulate Dense-Core Vesicle Maturation.” Neuron 82(1):167–80. doi: 10.1016/j.neuron.2014.02.017. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
