This is a preprint.
TDRD3 functions as a selective autophagy receptor with dual roles in autophagy and modulation of stress granule stability
- PMID: 39345463
- PMCID: PMC11430058
- DOI: 10.1101/2024.09.22.614367
TDRD3 functions as a selective autophagy receptor with dual roles in autophagy and modulation of stress granule stability
Abstract
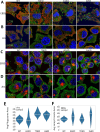
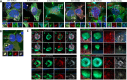
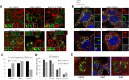
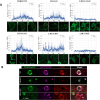
Tudor Domain Containing 3 (TDRD3) is a methylarginine-reader protein that functions as a scaffold in the nucleus facilitating transcription, however TDRD3 is also recruited to stress granules (SGs) during the Integrated Stress Response (ISR) although its function therein remains largely unknown. We previously showed that TDRD3 is a novel antiviral restriction factor that is cleaved by virus 2A protease, and plays complex modulatory roles in both interferon and inflammatory signaling during stress and enterovirus infections. Here we have found that TDRD3 contains structural motifs similar to known selective autophagy receptors such as p62/SQSTM1, sharing ubiquitin associated domains (UBA) and LC3 interacting regions (LIR) that anchor cargo destined for autophagosomes to activated LC3 protein coating autophagosome membranes. This is of interest since enteroviruses hijack autophagy machinery to facilitate formation of viral replication factories, virus assembly and egress from the infected cell. Here we explored possible roles of TDRD3 in autophagy, hypothesizing that TDRD3 may function as a specialized selective autophagy receptor. We found that KO of TDRD3 in HeLa cells significantly reduces starvation induced autophagy, while its reintroduction restores it in a dose-dependent manner. Autophagy receptors are degraded during autophagy and expression levels decrease during this time. We found that TDRD3 levels decrease to the same extent as the autophagy receptor p62/SQSTM1 during autophagy, indicating autophagy-targeted turnover in that role. Knockout of TDRD3 or G3BP1 did not make significant changes in overall cell localization of LC3B or p62/SQSTM1, but did result in greater concentration of Lamp2 phagosome marker for phagosomes and phagolysosomes. To test the potential roles of TDRD3 in autophagic processes, we created a series of deletion mutants of TDRD3 lacking either UBA domain or the various LIR motifs that are predicted to interact with LC3B. Microscopic examination of starved cells expressing these variants of TDRD3 showed ΔLIR-TDRD3 had defects in colocalization with LC3B or Lamp2. Further, super resolution microscopy revealed ring structures with TDRD3 interfacing with p62/SQSTM1. In examination of arsenite induced stress granules we found recruitment of TDRD3 variants disrupted normally tight SG condensation, altered the decay rate of SGs upon release from stress and the kinetics of SG formation. We found evidence that the LIR3 motif on TDRD3 is involved in TDRD3 interaction with LC3B in coIP experiments, colocalization studies, and that this motif plays a key role in TDRD3 recruitment to SGs and SG resolution. Overall, these data support a functional role of TDRD3 in selective autophagy in a mode similar to p62/SQSTM1, with specific roles in SG stability and turnover. Enterovirus cleavage of TDRD3 likely affects both antiviral and autophagic responses that the virus controls for replication.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist
Figures



References
-
- Linder B. et al. Tdrd3 is a novel stress granule-associated protein interacting with the Fragile-X syndrome protein FMRP. Human Molecular Genetics 17, 3236–3246 (2008). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
