This is a preprint.
CelloType: A Unified Model for Segmentation and Classification of Tissue Images
- PMID: 39345491
- PMCID: PMC11429831
- DOI: 10.1101/2024.09.15.613139
CelloType: A Unified Model for Segmentation and Classification of Tissue Images
Update in
-
CelloType: a unified model for segmentation and classification of tissue images.Nat Methods. 2025 Feb;22(2):348-357. doi: 10.1038/s41592-024-02513-1. Epub 2024 Nov 22. Nat Methods. 2025. PMID: 39578628 Free PMC article.
Abstract
Cell segmentation and classification are critical tasks in spatial omics data analysis. We introduce CelloType, an end-to-end model designed for cell segmentation and classification of biomedical microscopy images. Unlike the traditional two-stage approach of segmentation followed by classification, CelloType adopts a multi-task learning approach that connects the segmentation and classification tasks and simultaneously boost the performance of both tasks. CelloType leverages Transformer-based deep learning techniques for enhanced accuracy of object detection, segmentation, and classification. It outperforms existing segmentation methods using ground-truths from public databases. In terms of classification, CelloType outperforms a baseline model comprised of state-of-the-art methods for individual tasks. Using multiplexed tissue images, we further demonstrate the utility of CelloType for multi-scale segmentation and classification of both cellular and non-cellular elements in a tissue. The enhanced accuracy and multi-task-learning ability of CelloType facilitate automated annotation of rapidly growing spatial omics data.
Conflict of interest statement
Competing interests The authors declare no competing interests.
Figures



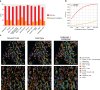
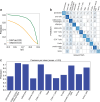
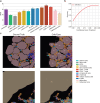
References
-
- Rozenblatt-Rosen O., Regev A., Oberdoerffer P., Nawy T., Hupalowska A., Rood J.E., Ashenberg O., Cerami E., Coffey R.J., Demir E., et al. (2020). The Human Tumor Atlas Network: Charting Tumor Transitions across Space and Time at Single-Cell Resolution. Cell 181, 236–249. 10.1016/j.cell.2020.03.053. - DOI - PMC - PubMed
-
- Greenwald N.F., Miller G., Moen E., Kong A., Kagel A., Dougherty T., Fullaway C.C., McIntosh B.J., Leow K.X., Schwartz M.S., et al. (2022). Whole-cell segmentation of tissue images with human-level performance using large-scale data annotation and deep learning. Nat Biotechnol 40, 555–565. 10.1038/s41587-021-01094-0. - DOI - PMC - PubMed
-
- He K., Zhang X., Ren S., Sun Ji. (2016). Deep Residual Learning for Image Recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), (IEEE), pp. 770–778.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
