This is a preprint.
RNA Polymerase II coordinates histone deacetylation at active promoters
- PMID: 39345547
- PMCID: PMC11429789
- DOI: 10.1101/2024.09.17.613553
RNA Polymerase II coordinates histone deacetylation at active promoters
Update in
-
RNA polymerase II coordinates histone deacetylation at active promoters.Sci Adv. 2025 Feb 7;11(6):eadt3037. doi: 10.1126/sciadv.adt3037. Epub 2025 Feb 5. Sci Adv. 2025. PMID: 39908363 Free PMC article.
Abstract
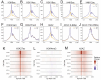
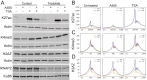
Nucleosomes at actively transcribed promoters have specific histone post-transcriptional modifications and histone variants. These features are thought to contribute to the formation and maintenance of a permissive chromatin environment. Recent reports have drawn conflicting conclusions about whether these histone modifications depend on transcription. We used triptolide to inhibit transcription initiation and degrade RNA Polymerase II and interrogated the effect on histone modifications. Transcription initiation was dispensable for de novo and steady-state histone acetylation at transcription start sites (TSSs) and enhancers. However, at steady state, blocking transcription initiation increased the levels of histone acetylation and H2AZ incorporation at active TSSs. These results demonstrate that deposition of specific histone modifications at TSSs is not dependent on transcription and that transcription limits the maintenance of these marks.
Conflict of interest statement
Competing interests: The authors declare that they have no competing interests.
Figures


References
-
- Talbert P. B., Henikoff S., The Yin and Yang of Histone Marks in Transcription. Annual Review of Genomics and Human Genetics 22, 147–170 (2021). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
